Short H2A histone variants are expressed in cancer
- PMID: 33473122
- PMCID: PMC7817690
- DOI: 10.1038/s41467-020-20707-x
Short H2A histone variants are expressed in cancer
Abstract
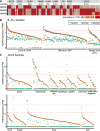
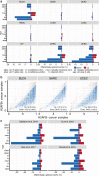
Short H2A (sH2A) histone variants are primarily expressed in the testes of placental mammals. Their incorporation into chromatin is associated with nucleosome destabilization and modulation of alternate splicing. Here, we show that sH2As innately possess features similar to recurrent oncohistone mutations associated with nucleosome instability. Through analyses of existing cancer genomics datasets, we find aberrant sH2A upregulation in a broad array of cancers, which manifest splicing patterns consistent with global nucleosome destabilization. We posit that short H2As are a class of "ready-made" oncohistones, whose inappropriate expression contributes to chromatin dysfunction in cancer.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

