CRISPR-Cas9 gene editing of hepatitis B virus in chronically infected humanized mice
- PMID: 33473359
- PMCID: PMC7803634
- DOI: 10.1016/j.omtm.2020.11.014
CRISPR-Cas9 gene editing of hepatitis B virus in chronically infected humanized mice
Abstract
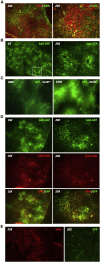
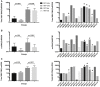
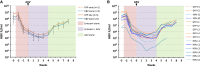
Chronic hepatitis B virus (HBV) infection is a major public health problem. New treatment approaches are needed because current treatments do not target covalently closed circular DNA (cccDNA), the template for HBV replication, and rarely clear the virus. We harnessed adeno-associated virus (AAV) vectors and CRISPR-Staphylococcus aureus (Sa)Cas9 to edit the HBV genome in liver-humanized FRG mice chronically infected with HBV and receiving entecavir. Gene editing was detected in livers of five of eight HBV-specific AAV-SaCas9-treated mice, but not control mice, and mice with detectable HBV gene editing showed higher levels of SaCas9 delivery to HBV+ human hepatocytes than those without gene editing. HBV-specific AAV-SaCas9 therapy significantly improved survival of human hepatocytes, showed a trend toward decreasing total liver HBV DNA and cccDNA, and was well tolerated. This work provides evidence for the feasibility and safety of in vivo gene editing for chronic HBV infections, and it suggests that with further optimization, this approach may offer a plausible way to treat or even cure chronic HBV infections.
Keywords: AAV; CRISPR/Cas9; Gene editing; HBV; Humanized mouse.
© 2020 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
-
- World Health Organization . 2019. Hepatitis B factsheet.https://www.who.int/en/news-room/fact-sheets/detail/hepatitis-b
-
- Zhu W., Xie K., Xu Y., Wang L., Chen K., Zhang L., Fang J. CRISPR/Cas9 produces anti-hepatitis B virus effect in hepatoma cells and transgenic mouse. Virus Res. 2016;217:125–132. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

