Transcriptional and functional characterization of neonatal circulating Innate Lymphoid Cells
- PMID: 33475258
- PMCID: PMC8133339
- DOI: 10.1002/sctm.20-0300
Transcriptional and functional characterization of neonatal circulating Innate Lymphoid Cells
Abstract
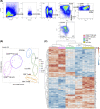
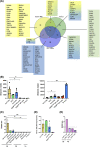
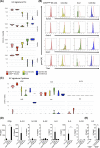
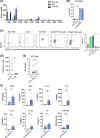
Innate lymphoid cells (ILCs), comprising ILC1, 2, and 3 subpopulations, play unique roles in maintaining microbiome homeostasis, mucosal tissue integrity, and control of inflammation. So far, their characterization is dominantly based on tissue-resident ILCs, whereas little information is available on circulating ILCs, in particular in newborns. In order to get a deeper understanding of neonatal innate immunity, we analyzed the transcriptomes and effector functions of cord blood (CB) ILCs. By RNAseq analysis, all ILC subsets could be clearly distinguished from each other. CB-derived ILCs were generally closer related to neonatal T than natural killer (NK) cells and several factors shared by all three ILC subsets such as CD28, CCR4, and SLAMF1 are commonly expressed by T cells but lacking in NK cells. Notably, CB ILCs exhibited a unique signature of DNA binding inhibitor (ID) transcription factors (TF) with high ID3 and low ID2 expression distinct from PB- or tonsil-derived ILCs. In vitro stimulation of sorted CB ILCs revealed distinct differences to tissue-resident ILCs in that ILC1-like and ILC3-like cells were nonresponsive to specific cytokine stimulation, indicating functional immaturity. However, CB ILC3-like cells expressed toll-like receptors TLR1 and TLR2 and upon stimulation with the TLR2:1 ligand Pam3 CSK4 , responded with significantly increased proliferation and cytokine secretion. Together, our data provide novel insights into neonatal ILC biology with a unique TF signature of CB ILCs possibly indicating a common developmental pathway and furthermore a role of CB ILC3-like cells in innate host defense.
Keywords: ID3; ILCs; TLR2:1; transcriptome; umbilical cord blood.
© 2021 The Authors. STEM CELLS TRANSLATIONAL MEDICINE published by Wiley Periodicals LLC on behalf of AlphaMed Press.
Conflict of interest statement
The authors declared no potential conflicts of interest.
Figures

References
-
- Spits H, Artis D, Colonna M, et al. Innate lymphoid cells ‐ a proposal for uniform nomenclature. Nat Rev Immunol. 2013;13(2):145‐149. - PubMed
-
- Vivier E, Artis D, Colonna M, et al. Innate lymphoid cells: 10 years on. Cell. 2018;174(5):1054‐1066. - PubMed
-
- Lim AI, Li Y, Lopez‐Lastra S, et al. Systemic human ILC precursors provide a substrate for tissue ILC differentiation. Cell. 2017;168(6):1086‐1100.e1010. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

