Inositol phosphates promote HIV-1 assembly and maturation to facilitate viral spread in human CD4+ T cells
- PMID: 33476323
- PMCID: PMC7853515
- DOI: 10.1371/journal.ppat.1009190
Inositol phosphates promote HIV-1 assembly and maturation to facilitate viral spread in human CD4+ T cells
Erratum in
-
Correction: Inositol phosphates promote HIV-1 assembly and maturation to facilitate viral spread in human CD4+ T cells.PLoS Pathog. 2021 Mar 2;17(3):e1009389. doi: 10.1371/journal.ppat.1009389. eCollection 2021 Mar. PLoS Pathog. 2021. PMID: 33651846 Free PMC article.
Abstract
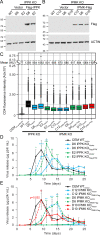
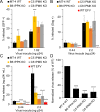
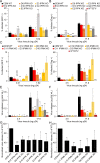
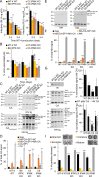
Gag polymerization with viral RNA at the plasma membrane initiates HIV-1 assembly. Assembly processes are inefficient in vitro but are stimulated by inositol (1,3,4,5,6) pentakisphosphate (IP5) and inositol hexakisphosphate (IP6) metabolites. Previous studies have shown that depletion of these inositol phosphate species from HEK293T cells reduced HIV-1 particle production but did not alter the infectivity of the resulting progeny virions. Moreover, HIV-1 substitutions bearing Gag/CA mutations ablating IP6 binding are noninfectious with destabilized viral cores. In this study, we analyzed the effects of cellular depletion of IP5 and IP6 on HIV-1 replication in T cells in which we disrupted the genes encoding the kinases required for IP6 generation, IP5 2-kinase (IPPK) and Inositol Polyphosphate Multikinase (IPMK). Knockout (KO) of IPPK from CEM and MT-4 cells depleted cellular IP6 in both T cell lines, and IPMK disruption reduced the levels of both IP5 and IP6. In the KO lines, HIV-1 spread was delayed relative to parental wild-type (WT) cells and was rescued by complementation. Virus release was decreased in all IPPK or IPMK KO lines relative to WT cells. Infected IPMK KO cells exhibited elevated levels of intracellular Gag protein, indicative of impaired particle assembly. IPMK KO compromised virus production to a greater extent than IPPK KO suggesting that IP5 promotes HIV-1 particle assembly in IPPK KO cells. HIV-1 particles released from infected IPPK or IPMK KO cells were less infectious than those from WT cells. These viruses exhibited partially cleaved Gag proteins, decreased virion-associated p24, and higher frequencies of aberrant particles, indicative of a maturation defect. Our data demonstrate that IP6 enhances the quantity and quality of virions produced from T cells, thereby preventing defects in HIV-1 replication.
Conflict of interest statement
No authors have competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

