SARS-CoV-2 variants with mutations at the S1/S2 cleavage site are generated in vitro during propagation in TMPRSS2-deficient cells
- PMID: 33476327
- PMCID: PMC7853460
- DOI: 10.1371/journal.ppat.1009233
SARS-CoV-2 variants with mutations at the S1/S2 cleavage site are generated in vitro during propagation in TMPRSS2-deficient cells
Abstract
The spike (S) protein of Severe Acute Respiratory Syndrome-Coronavirus-2 (SARS-CoV-2) binds to a host cell receptor which facilitates viral entry. A polybasic motif detected at the cleavage site of the S protein has been shown to broaden the cell tropism and transmissibility of the virus. Here we examine the properties of SARS-CoV-2 variants with mutations at the S protein cleavage site that undergo inefficient proteolytic cleavage. Virus variants with S gene mutations generated smaller plaques and exhibited a more limited range of cell tropism compared to the wild-type strain. These alterations were shown to result from their inability to utilize the entry pathway involving direct fusion mediated by the host type II transmembrane serine protease, TMPRSS2. Notably, viruses with S gene mutations emerged rapidly and became the dominant SARS-CoV-2 variants in TMPRSS2-deficient cells including Vero cells. Our study demonstrated that the S protein polybasic cleavage motif is a critical factor underlying SARS-CoV-2 entry and cell tropism. As such, researchers should be alert to the possibility of de novo S gene mutations emerging in tissue-culture propagated virus strains.
Conflict of interest statement
I have read the journal's policy and the authors of this manuscript have the following competing interests: K.U., A.S., S.T., and T.S. are employees of Shionogi & Co., Ltd. Other authors have declared that no competing interests exist.
Figures
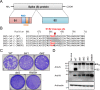

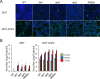
References
-
- (WHO) WHO. Coronavirus disease (COVID-19) pandemic 2020 [cited 2020 9th November]. Available from: https://www.who.int/emergencies/diseases/novel-coronavirus-2019.
-
- Naqvi AAT, Fatima K, Mohammad T, Fatima U, Singh IK, Singh A, et al. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: Structural genomics approach. Biochim Biophys Acta Mol Basis Dis. 2020;1866(10):165878 Epub 2020/06/13. 10.1016/j.bbadis.2020.165878 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

