RPA complexes in Caenorhabditis elegans meiosis; unique roles in replication, meiotic recombination and apoptosis
- PMID: 33476370
- PMCID: PMC7913698
- DOI: 10.1093/nar/gkaa1293
RPA complexes in Caenorhabditis elegans meiosis; unique roles in replication, meiotic recombination and apoptosis
Erratum in
-
Correction to 'RPA complexes in Caenorhabditis elegans meiosis; unique roles in replication, meiotic recombination and apoptosis'.Nucleic Acids Res. 2021 Sep 7;49(15):9003. doi: 10.1093/nar/gkab631. Nucleic Acids Res. 2021. PMID: 34370030 Free PMC article. No abstract available.
Abstract
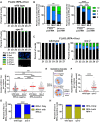
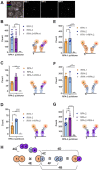
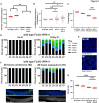
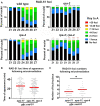
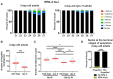
Replication Protein A (RPA) is a critical complex that acts in replication and promotes homologous recombination by allowing recombinase recruitment to processed DSB ends. Most organisms possess three RPA subunits (RPA1, RPA2, RPA3) that form a trimeric complex critical for viability. The Caenorhabditis elegans genome encodes RPA-1, RPA-2 and an RPA-2 paralog RPA-4. In our analysis, we determined that RPA-2 is critical for germline replication and normal repair of meiotic DSBs. Interestingly, RPA-1 but not RPA-2 is essential for somatic replication, in contrast to other organisms that require both subunits. Six different hetero- and homodimeric complexes containing permutations of RPA-1, RPA-2 and RPA-4 can be detected in whole animal extracts. Our in vivo studies indicate that RPA-1/4 dimer is less abundant in the nucleus and its formation is inhibited by RPA-2. While RPA-4 does not participate in replication or recombination, we find that RPA-4 inhibits RAD-51 filament formation and promotes apoptosis of a subset of damaged nuclei. Altogether these findings point to sub-functionalization and antagonistic roles of RPA complexes in C. elegans.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- Treuner K., Ramsperger U., Knippers R.. Replication protein A induces the unwinding of long double-stranded DNA regions. J. Mol. Biol. 1996; 259:104–112. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

