A Legionella effector ADP-ribosyltransferase inactivates glutamate dehydrogenase
- PMID: 33476647
- PMCID: PMC7949102
- DOI: 10.1016/j.jbc.2021.100301
A Legionella effector ADP-ribosyltransferase inactivates glutamate dehydrogenase
Abstract
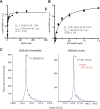
ADP-ribosyltransferases (ARTs) are a widespread superfamily of enzymes frequently employed in pathogenic strategies of bacteria. Legionella pneumophila, the causative agent of a severe form of pneumonia known as Legionnaire's disease, has acquired over 330 translocated effectors that showcase remarkable biochemical and structural diversity. However, the ART effectors that influence L. pneumophila have not been well defined. Here, we took a bioinformatic approach to search the Legionella effector repertoire for additional divergent members of the ART superfamily and identified an ART domain in Legionella pneumophila gene0181, which we hereafter refer to as Legionella ADP-Ribosyltransferase 1 (Lart1) (Legionella ART 1). We show that L. pneumophila Lart1 targets a specific class of 120-kDa NAD+-dependent glutamate dehydrogenase (GDH) enzymes found in fungi and protists, including many natural hosts of Legionella. Lart1 targets a conserved arginine residue in the NAD+-binding pocket of GDH, thereby blocking oxidative deamination of glutamate. Therefore, Lart1 could be the first example of a Legionella effector which directly targets a host metabolic enzyme during infection.
Keywords: ADP-ribosylation; bacterial pathogenesis; bioinformatics; cell metabolism; enzyme kinetics; glutamate; glutamate dehydrogenase; infectious disease.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures




References
-
- Gomez-Valero L., Rusniok C., Carson D., Mondino S., Perez-Cobas A.E., Rolando M., Pasricha S., Reuter S., Demirtas J., Crumbach J., Descorps-Declere S., Hartland E.L., Jarraud S., Dougan G., Schroeder G.N. More than 18,000 effectors in the Legionella genus genome provide multiple, independent combinations for replication in human cells. Proc. Natl. Acad. Sci. U. S. A. 2019;116:2265–2273. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

