The 442th amino acid residue of the spike protein is critical for the adaptation to bat hosts for SARS-related coronaviruses
- PMID: 33476695
- PMCID: PMC7813513
- DOI: 10.1016/j.virusres.2021.198307
The 442th amino acid residue of the spike protein is critical for the adaptation to bat hosts for SARS-related coronaviruses
Abstract
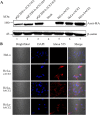
Bats carry diverse severe acute respiratory syndrome-related coronaviruses (SARSr-CoVs). The suspected interspecies transmission of SARSr-CoVs from bats to humans has caused two severe CoV pandemics, the SARS pandemic in 2003 and the recent COVID-19 pandemic. The receptor utilization of SARSr-CoV plays the key role in determining the host range and the interspecies transmission ability of the virus. Both SARS-CoV and SARS-CoV-2 use angiotensin-converting enzyme 2 (ACE2) as their receptor. Previous studies showed that WIV1 strain, the first living coronavirus isolated from bat using ACE2 as its receptor, is the prototype of SARS-CoV. The receptor-binding domain (RBD) in the spike protein (S) of SARS-CoV and WIV1 is responsible for ACE2 binding and medicates the viral entry. Comparing to SARS-CoV, WIV1 has three distinct amino acid residues (442, 472, and 487) in its RBD. This study aimed at exploring whether these three residues could alter the receptor utilization of SARSr-CoVs. We replaced the three residues in SARS-CoV (BJ01 strain) S with their counterparts in WIV1 S, and then evaluated the change of their utilization of bat, civet, and human ACE2s using a lentivirus-based pseudovirus infection system. To further validate the S-ACE2 interactions, the binding affinity between the RBDs of these S proteins and the three ACE2s were verified by flow cytometry. The results showed that the single amino acid substitution Y442S in the RBD of BJ01 S enhanced its utilization of bat ACE2 and its binding affinity to bat ACE2. On the contrary, the reverse substitution in WIV1 S (S442Y) significantly attenuated the pseudovirus utilization of bat, civet and human ACE2s for cell entry, and reduced its binding affinity with the three ACE2s. These results suggest that the S442 is critical for WIV1 adapting to bats as its natural hosts. These findings will enhance our understanding of host adaptations and cross-species infections of coronaviruses, contributing to the prediction and prevention of coronavirus epidemics.
Keywords: ACE2; Receptor binding domain; Receptor utilization; SARS-CoV; SARSr-CoV; Spike.
Copyright © 2021 Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors report no declarations of interest.
Figures


Similar articles
-
Evolutionary Arms Race between Virus and Host Drives Genetic Diversity in Bat Severe Acute Respiratory Syndrome-Related Coronavirus Spike Genes.J Virol. 2020 Sep 29;94(20):e00902-20. doi: 10.1128/JVI.00902-20. Print 2020 Sep 29. J Virol. 2020. PMID: 32699095 Free PMC article.
-
Structural determinants of spike infectivity in bat SARS-like coronaviruses RsSHC014 and WIV1.J Virol. 2024 Aug 20;98(8):e0034224. doi: 10.1128/jvi.00342-24. Epub 2024 Jul 19. J Virol. 2024. PMID: 39028202 Free PMC article.
-
Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor.Nature. 2013 Nov 28;503(7477):535-8. doi: 10.1038/nature12711. Epub 2013 Oct 30. Nature. 2013. PMID: 24172901 Free PMC article.
-
Inhibition of S-protein RBD and hACE2 Interaction for Control of SARSCoV- 2 Infection (COVID-19).Mini Rev Med Chem. 2021;21(6):689-703. doi: 10.2174/1389557520666201117111259. Mini Rev Med Chem. 2021. PMID: 33208074 Review.
-
Molecular epidemiology, evolution and phylogeny of SARS coronavirus.Infect Genet Evol. 2019 Jul;71:21-30. doi: 10.1016/j.meegid.2019.03.001. Epub 2019 Mar 4. Infect Genet Evol. 2019. PMID: 30844511 Free PMC article. Review.
Cited by
-
Binding affinity between coronavirus spike protein and human ACE2 receptor.Comput Struct Biotechnol J. 2024 Jan 17;23:759-770. doi: 10.1016/j.csbj.2024.01.009. eCollection 2024 Dec. Comput Struct Biotechnol J. 2024. PMID: 38304547 Free PMC article. Review.
-
Key mutations in the spike protein of SARS-CoV-2 affecting neutralization resistance and viral internalization.J Med Virol. 2023 Jan;95(1):e28407. doi: 10.1002/jmv.28407. J Med Virol. 2023. PMID: 36519597 Free PMC article.
References
-
- Becker M.M., Graham R.L., Donaldson E.F., Rockx B., Sims A.C., Sheahan T.P., Pickles R.J., Davide C., Johnston R.E., Baric R.S., Denison M.R. Synthetic recombinant bat SARS-like coronavirus is infectious in cultured cells and in mice. Proc. Natl. Acad. Sci. U. S. A. 2008;105(50):19944–19949. - PMC - PubMed
-
- Bertram S., Glowacka I., Muller M.A., Lavender H., Gnirss K., Nehlmeier I., Niemeyer D., He Y., Simmons G., Drosten C., Soilleux E.J., Jahn O., Steffen I., Pohlmann S. Cleavage and activation of the severe acute respiratory syndrome coronavirus spike protein by human airway trypsin-like protease. J. Virol. 2011;85(24):13363–13372. - PMC - PubMed
-
- Burgin C.J., Colella J.P., Kahn P.L., Upham N.S. How many species of mammals are there? J. Mammal. 2018;99(1):1–14.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

