LRIG1, a regulator of stem cell quiescence and a pleiotropic feedback tumor suppressor
- PMID: 33476721
- PMCID: PMC8286266
- DOI: 10.1016/j.semcancer.2020.12.016
LRIG1, a regulator of stem cell quiescence and a pleiotropic feedback tumor suppressor
Abstract
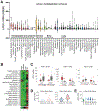
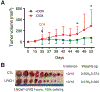
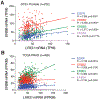
LRIG1, leucine-rich repeats and immunoglobulin-like domains protein 1, was discovered more than 20 years ago and has been shown to be downregulated or lost, and to function as a tumor suppressor in several cancers. Another well-reported biological function of LRIG1 is to regulate and help enforce the quiescence of adult stem cells (SCs). In both contexts, LRIG1 regulates SC quiescence and represses tumor growth via, primarily, antagonizing the expression and activities of ERBB and other receptor tyrosine kinases (RTKs). We have recently reported that in treatment-naïve human prostate cancer (PCa), LRIG1 is primarily regulated by androgen receptor (AR) and is prominently overexpressed. In castration-resistant PCa (CRPC), both LRIG1 and AR expression becomes heterogeneous and, frequently, discordant. Importantly, in both androgen-dependent PCa and CRPC models, LRIG1 exhibits tumor-suppressive functions. Moreover, LRIG1 induction inhibits the growth of pre-established AR+ and AR- PCa. Here, upon a brief introduction of the LRIG1 and the LRIG family, we provide an updated overview on LRIG1 functions in regulating SC quiescence and repressing tumor development. We further highlight the expression, regulation and functions of LRIG1 in treatment-naïve PCa and CRPC. We conclude by offering the perspectives of identifying novel cancer-specific LRIG1-interacting signaling partners and developing LRIG1-based anti-cancer therapeutics and diagnostic/prognostic biomarkers.
Keywords: Cancer stem cells; LRIG1; Prostate cancer; Stem cells; Tumor suppressor.
Copyright © 2021 The Authors. Published by Elsevier Ltd.. All rights reserved.
Figures




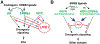
References
-
- Suzuki Y, Sato N, Tohyama M, et al., cDNA doning of a novel membrane glycoprotein that is expressed specifically in glial cells in the mouse brain. LIG-1, a protein with leucine-rich repeats and immunoglobulin-like domains, J. Biol. Chem 271 (37) (1996) 22522–22527, 10.1074/jbc.271.37.22522. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

