Novel nucleocytoplasmic protein O-fucosylation by SPINDLY regulates diverse developmental processes in plants
- PMID: 33476897
- PMCID: PMC8222059
- DOI: 10.1016/j.sbi.2020.12.013
Novel nucleocytoplasmic protein O-fucosylation by SPINDLY regulates diverse developmental processes in plants
Abstract
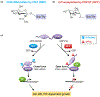
In metazoans, protein O-fucosylation of Ser/Thr residues was only found in secreted or cell surface proteins, and this post-translational modification is catalyzed by ER-localized protein O-fucosyltransferases (POFUTs) in the GT65 family. Recently, a novel nucleocytoplasmic POFUT, SPINDLY (SPY), was identified in the reference plant Arabidopsis thaliana to modify nuclear transcription regulators DELLAs, revealing a new regulatory mechanism for gene expression. The paralog of AtSPY, SECRET AGENT (SEC), is an O-link-N-acetylglucosamine (GlcNAc) transferase (OGT), which O-GlcNAcylates Ser/Thr residues of target proteins. Both AtSPY and AtSEC are tetratricopeptide repeat-domain-containing glycosyltransferases in the GT41 family. The discovery that AtSPY is a POFUT clarified decades of miss-classification of AtSPY as an OGT. SPY and SEC play pleiotropic roles in plant development, and the interactions between SPY and SEC are complex. SPY-like genes are conserved in diverse organisms, except in fungi and metazoans, suggesting that O-fucosylation is a common mechanism in modulating intracellular protein functions.
Copyright © 2021 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Conflict of Interest
No conflict of interest exists
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

