HIV Capsid and Integration Targeting
- PMID: 33477441
- PMCID: PMC7830116
- DOI: 10.3390/v13010125
HIV Capsid and Integration Targeting
Abstract
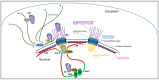
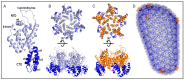
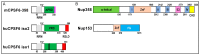
Integration of retroviral reverse transcripts into the chromosomes of the cells that they infect is required for efficient viral gene expression and the inheritance of viral genomes to daughter cells. Before integration can occur, retroviral reverse transcription complexes (RTCs) must access the nuclear environment where the chromosomes reside. Retroviral integration is non-random, with different types of virus-host interactions impacting where in the host chromatin integration takes place. Lentiviruses such as HIV efficiently infect interphase cells because their RTCs have evolved to usurp cellular nuclear import transport mechanisms, and research over the past decade has revealed specific interactions between the HIV capsid protein and nucleoporin (Nup) proteins such as Nup358 and Nup153. The interaction of HIV capsid with cleavage and polyadenylation specificity factor 6 (CPSF6), which is a component of the cellular cleavage and polyadenylation complex, helps to dictate nuclear import as well as post-nuclear RTC invasion. In the absence of the capsid-CPSF6 interaction, RTCs are precluded from reaching nuclear speckles and gene-rich regions of chromatin known as speckle-associated domains, and instead mis-target lamina-associated domains out at the nuclear periphery. Highlighting this area of research, small molecules that inhibit capsid-host interactions important for integration site targeting are highly potent antiviral compounds.
Keywords: CPSF6; HIV; antiviral inhibitor; capsid; integration; integration targeting; nuclear import.
Conflict of interest statement
A.N.E. has been compensated by ViiV Healthcare Co. for consulting on unrelated work.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

