Modeling the Influenza A NP-vRNA-Polymerase Complex in Atomic Detail
- PMID: 33477938
- PMCID: PMC7833383
- DOI: 10.3390/biom11010124
Modeling the Influenza A NP-vRNA-Polymerase Complex in Atomic Detail
Abstract
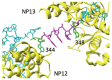
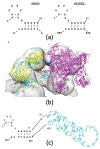
Seasonal flu is an acute respiratory disease that exacts a massive toll on human populations, healthcare systems and economies. The disease is caused by an enveloped Influenza virus containing eight ribonucleoprotein (RNP) complexes. Each RNP incorporates multiple copies of nucleoprotein (NP), a fragment of the viral genome (vRNA), and a viral RNA-dependent RNA polymerase (POL), and is responsible for packaging the viral genome and performing critical functions including replication and transcription. A complete model of an Influenza RNP in atomic detail can elucidate the structural basis for viral genome functions, and identify potential targets for viral therapeutics. In this work we construct a model of a complete Influenza A RNP complex in atomic detail using multiple sources of structural and sequence information and a series of homology-modeling techniques, including a motif-matching fragment assembly method. Our final model provides a rationale for experimentally-observed changes to viral polymerase activity in numerous mutational assays. Further, our model reveals specific interactions between the three primary structural components of the RNP, including potential targets for blocking POL-binding to the NP-vRNA complex. The methods developed in this work open the possibility of elucidating other functionally-relevant atomic-scale interactions in additional RNP structures and other biomolecular complexes.
Keywords: Influenza A; RNP; homology modeling; viral genome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Iuliano A.D., Roguski K.M., Chang H.H., Muscatello D.J., Palekar R., Tempia S., Cohen C., Gran J.M., Schanzer D., Cowling B.J., et al. Estimates of global seasonal influenza-associated respiratory mortality: A modelling study. Lancet. 2018;391:1285–1300. doi: 10.1016/S0140-6736(17)33293-2. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

