Fecal Microbiota Transplant from Human to Mice Gives Insights into the Role of the Gut Microbiota in Non-Alcoholic Fatty Liver Disease (NAFLD)
- PMID: 33477939
- PMCID: PMC7833443
- DOI: 10.3390/microorganisms9010199
Fecal Microbiota Transplant from Human to Mice Gives Insights into the Role of the Gut Microbiota in Non-Alcoholic Fatty Liver Disease (NAFLD)
Abstract
Non-alcoholic fatty liver diseases (NAFLD) are associated with changes in the composition and metabolic activities of the gut microbiota. However, the causal role played by the gut microbiota in individual susceptibility to NAFLD and particularly at its early stage is still unclear. In this context, we transplanted the microbiota from a patient with fatty liver (NAFL) and from a healthy individual to two groups of mice. We first showed that the microbiota composition in recipient mice resembled the microbiota composition of their respective human donor. Following administration of a high-fructose, high-fat diet, mice that received the human NAFL microbiota (NAFLR) gained more weight and had a higher liver triglycerides level and higher plasma LDL cholesterol than mice that received the human healthy microbiota (HR). Metabolomic analyses revealed that it was associated with lower and higher plasma levels of glycine and 3-Indolepropionic acid in NAFLR mice, respectively. Moreover, several bacterial genera and OTUs were identified as differently represented in the NAFLR and HR microbiota and therefore potentially responsible for the different phenotypes observed. Altogether, our results confirm that the gut bacteria play a role in obesity and steatosis development and that targeting the gut microbiota may be a preventive or therapeutic strategy in NAFLD management.
Keywords: NAFL; NAFLD; gut microbiome; health; high-fat diet; high-fructose; human; liver.
Conflict of interest statement
J.-M.P., W.F., F.S. and L.C. are employees of the Enterome Company. The other authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures
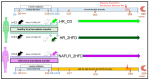




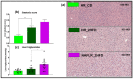





References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

