Genome-based engineering of ligninolytic enzymes in fungi
- PMID: 33478513
- PMCID: PMC7819241
- DOI: 10.1186/s12934-021-01510-9
Genome-based engineering of ligninolytic enzymes in fungi
Abstract
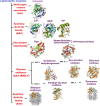
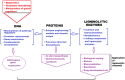
Background: Many fungi grow as saprobic organisms and obtain nutrients from a wide range of dead organic materials. Among saprobes, fungal species that grow on wood or in polluted environments have evolved prolific mechanisms for the production of degrading compounds, such as ligninolytic enzymes. These enzymes include arrays of intense redox-potential oxidoreductase, such as laccase, catalase, and peroxidases. The ability to produce ligninolytic enzymes makes a variety of fungal species suitable for application in many industries, including the production of biofuels and antibiotics, bioremediation, and biomedical application as biosensors. However, fungal ligninolytic enzymes are produced naturally in small quantities that may not meet the industrial or market demands. Over the last decade, combined synthetic biology and computational designs have yielded significant results in enhancing the synthesis of natural compounds in fungi. In this review, we gave insights into different protein engineering methods, including rational, semi-rational, and directed evolution approaches that have been employed to enhance the production of some important ligninolytic enzymes in fungi. We described the role of metabolic pathway engineering to optimize the synthesis of chemical compounds of interest in various fields. We highlighted synthetic biology novel techniques for biosynthetic gene cluster (BGC) activation in fungo and heterologous reconstruction of BGC in microbial cells. We also discussed in detail some recombinant ligninolytic enzymes that have been successfully enhanced and expressed in different heterologous hosts. Finally, we described recent advance in CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats)-Cas (CRISPR associated) protein systems as the most promising biotechnology for large-scale production of ligninolytic enzymes.
Short conclusion: Aggregation, expression, and regulation of ligninolytic enzymes in fungi require very complex procedures with many interfering factors. Synthetic and computational biology strategies, as explained in this review, are powerful tools that can be combined to solve these puzzles. These integrated strategies can lead to the production of enzymes with special abilities, such as wide substrate specifications, thermo-stability, tolerance to long time storage, and stability in different substrate conditions, such as pH and nutrients.
Keywords: Biosynthetic pathways; CRISPR-cas; Fungal secretome; Fungi; Gene editing; Heterologous protein expression; Ligninolytic enzymes; Synthetic promoters; Transcription activation.
Conflict of interest statement
Authors declare no competing interest on this manuscript.
Figures


References
-
- Blanchette RA, Krueger EW, Haight JE, Akhtar M, Akin DE. Cell wall alterations in loblolly pine wood decayed by the white-rot fungus Ceriporiopsis subvermispora. J Biotechnol. 1997;53:203–213. doi: 10.1016/S0168-1656(97)01674-X. - DOI
-
- Asemoloye MD, Tosi S, Daccò C, Wang X, Xu S, Marchisio MA, Gao W, Jonathan SG, Pecoraro L. Hydrocarbon degradation and enzyme activities of Aspergillus oryzae and Mucor irregularis isolated from Nigerian crude oil-polluted sites. Microorganisms. 2020;8(12):1912. doi: 10.3390/microorganisms8121912. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

