A Scalable, Easy-to-Deploy Protocol for Cas13-Based Detection of SARS-CoV-2 Genetic Material
- PMID: 33478979
- PMCID: PMC8092748
- DOI: 10.1128/JCM.02402-20
A Scalable, Easy-to-Deploy Protocol for Cas13-Based Detection of SARS-CoV-2 Genetic Material
Abstract
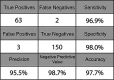
The COVID-19 pandemic has created massive demand for widespread, distributed tools for detecting SARS-CoV-2 genetic material. The hurdles to scalable testing include reagent and instrument accessibility, availability of highly trained personnel, and large upfront investment. Here, we showcase an orthogonal pipeline we call CREST (Cas13-based, rugged, equitable, scalable testing) that addresses some of these hurdles. Specifically, CREST pairs commonplace and reliable biochemical methods (PCR) with low-cost instrumentation, without sacrificing detection sensitivity. By taking advantage of simple fluorescence visualizers, CREST allows a binary interpretation of results. CREST may provide a point-of-care solution to increase the distribution of COVID-19 surveillance.
Keywords: COVID-19; CRISPR; Cas13; SARS-CoV-2; point of care; testing.
Copyright © 2021 Rauch et al.
Figures



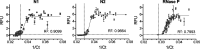
References
-
- Mizumoto K, Kagaya K, Zarebski A, medRxiv GC. 2020. Estimating the asymptomatic ratio of 2019 novel coronavirus onboard the Princess Cruises ship, 2020. medrxiv.org.
-
- Hu Z, Song C, Xu C, Jin G, Chen Y, Xu X, Ma H, Chen W, Lin Y, Zheng Y, Wang J, Hu Z, Yi Y, Shen H. 2020. Clinical characteristics of 24 asymptomatic infections with COVID-19 screened among close contacts in Nanjing, China. Sci China Life Sci 63:706–711. doi: 10.1007/s11427-020-1661-4. - DOI - PMC - PubMed
-
- Wölfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Müller MA, Niemeyer D, Jones TC, Vollmar P, Rothe C, Hoelscher M, Bleicker T, Brünink S, Schneider J, Ehmann R, Zwirglmaier K, Drosten C, Wendtner C. 2020. Virological assessment of hospitalized patients with COVID-2019. Nature 581:465–469. doi: 10.1038/s41586-020-2196-x. - DOI - PubMed
-
- He X, Lau EHY, Wu P, Deng X, Wang J, Hao X, Lau YC, Wong JY, Guan Y, Tan X, Mo X, Chen Y, Liao B, Chen W, Hu F, Zhang Q, Zhong M, Wu Y, Zhao L, Zhang F, Cowling BJ, Li F, Leung GM. 2020. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat Med 26:672–675. doi: 10.1038/s41591-020-0869-5. - DOI - PubMed
-
- Verity R, Okell LC, Dorigatti I, Winskill P, Whittaker C, Imai N, Cuomo-Dannenburg G, Thompson H, Walker PGT, Fu H, Dighe A, Griffin JT, Baguelin M, Bhatia S, Boonyasiri A, Cori A, Cucunubá Z, FitzJohn R, Gaythorpe K, Green W, Hamlet A, Hinsley W, Laydon D, Nedjati-Gilani G, Riley S, van Elsland S, Volz E, Wang H, Wang Y, Xi X, Donnelly CA, Ghani AC, Ferguson NM. 2020. Estimates of the severity of coronavirus disease 2019: a model-based analysis. Lancet Infect Dis 20:669–677. doi: 10.1016/S1473-3099(20)30243-7. - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

