Dosage-sensitive functions in embryonic development drove the survival of genes on sex-specific chromosomes in snakes, birds, and mammals
- PMID: 33479023
- PMCID: PMC7849413
- DOI: 10.1101/gr.268516.120
Dosage-sensitive functions in embryonic development drove the survival of genes on sex-specific chromosomes in snakes, birds, and mammals
Abstract
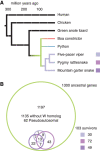
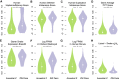
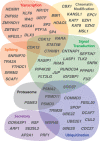
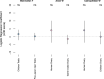
Different ancestral autosomes independently evolved into sex chromosomes in snakes, birds, and mammals. In snakes and birds, females are ZW and males are ZZ; in mammals, females are XX and males are XY. Although X and Z Chromosomes retain nearly all ancestral genes, sex-specific W and Y Chromosomes suffered extensive genetic decay. In both birds and mammals, the genes that survived on sex-specific chromosomes are enriched for broadly expressed, dosage-sensitive regulators of gene expression, subject to strong purifying selection. To gain deeper insight into the processes that govern survival on sex-specific chromosomes, we carried out a meta-analysis of survival across 41 species-three snakes, 24 birds, and 14 mammals-doubling the number of ancestral genes under investigation and increasing our power to detect enrichments among survivors relative to nonsurvivors. Of 2564 ancestral genes, representing an eighth of the ancestral amniote genome, only 324 survive on present-day sex-specific chromosomes. Survivors are enriched for dosage-sensitive developmental processes, particularly development of neural crest-derived structures, such as the face. However, there was no enrichment for expression in sex-specific tissues, involvement in sex determination or gonadogenesis pathways, or conserved sex-biased expression. Broad expression and dosage sensitivity contributed independently to gene survival, suggesting that pleiotropy imposes additional constraints on the evolution of dosage compensation. We propose that maintaining the viability of the heterogametic sex drove gene survival on amniote sex-specific chromosomes, and that subtle modulation of the expression of survivor genes and their autosomal orthologs has disproportionately large effects on development and disease.
© 2021 Bellott and Page; Published by Cold Spring Harbor Laboratory Press.
Figures

References
-
- Ayers KL, Davidson NM, Demiyah D, Roeszler KN, Grützner F, Sinclair AH, Oshlack A, Smith CA. 2013. RNA sequencing reveals sexually dimorphic gene expression before gonadal differentiation in chicken and allows comprehensive annotation of the W-chromosome. Genome Biol 14: R26 10.1186/gb-2013-14-3-r26 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
