Dynamic association of human Ebp1 with the ribosome
- PMID: 33479117
- PMCID: PMC7962488
- DOI: 10.1261/rna.077602.120
Dynamic association of human Ebp1 with the ribosome
Abstract
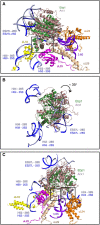
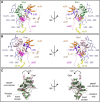
Ribosomes are the macromolecular machines at the heart of protein synthesis; however, their function can be modulated by a variety of additional protein factors that directly interact with them. Here, we report the cryo-EM structure of human Ebp1 (p48 isoform) bound to the human 80S ribosome at 3.3 Å resolution. Ebp1 binds in the vicinity of the peptide exit tunnel on the 80S ribosome, and this binding is enhanced upon puromycin-mediated translational inhibition. The association of Ebp1 with the 80S ribosome centers around its interaction with ribosomal proteins eL19 and uL23 and the 28S rRNA. Further analysis of the Ebp1-ribosome complex suggests that Ebp1 can rotate around its insert domain, which may enable it to assume a wide range of conformations while maintaining its interaction with the ribosome. Structurally, Ebp1 shares homology with the methionine aminopeptidase 2 family of enzymes; therefore, this inherent flexibility may also be conserved.
Keywords: Ebp1; ribosome; single-particle cryo-EM.
© 2021 Bhaskar et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures

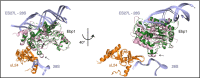
References
-
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, et al. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221. 10.1107/S0907444909052925 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
