Structural basis for transcription complex disruption by the Mfd translocase
- PMID: 33480355
- PMCID: PMC7864632
- DOI: 10.7554/eLife.62117
Structural basis for transcription complex disruption by the Mfd translocase
Abstract
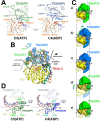
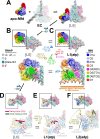
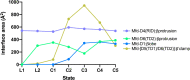
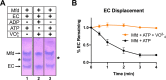
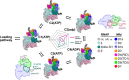
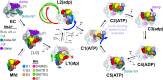
Transcription-coupled repair (TCR) is a sub-pathway of nucleotide excision repair (NER) that preferentially removes lesions from the template-strand (t-strand) that stall RNA polymerase (RNAP) elongation complexes (ECs). Mfd mediates TCR in bacteria by removing the stalled RNAP concealing the lesion and recruiting Uvr(A)BC. We used cryo-electron microscopy to visualize Mfd engaging with a stalled EC and attempting to dislodge the RNAP. We visualized seven distinct Mfd-EC complexes in both ATP and ADP-bound states. The structures explain how Mfd is remodeled from its repressed conformation, how the UvrA-interacting surface of Mfd is hidden during most of the remodeling process to prevent premature engagement with the NER pathway, how Mfd alters the RNAP conformation to facilitate disassembly, and how Mfd forms a processive translocation complex after dislodging the RNAP. Our results reveal an elaborate mechanism for how Mfd kinetically discriminates paused from stalled ECs and disassembles stalled ECs to initiate TCR.
Keywords: E. coli; Mfd; RNA polymerase; cryo-electron microscopy; molecular biophysics; structural biology; transcription repair coupling factor; transcription-coupled repair.
© 2021, Kang et al.
Conflict of interest statement
JK, EL, JC, PO, JB, BC, EC, SD No competing interests declared
Figures



References
-
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica Section D Biological Crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

