DNA topoisomerases: Advances in understanding of cellular roles and multi-protein complexes via structure-function analysis
- PMID: 33480441
- PMCID: PMC7614492
- DOI: 10.1002/bies.202000286
DNA topoisomerases: Advances in understanding of cellular roles and multi-protein complexes via structure-function analysis
Abstract
DNA topoisomerases, capable of manipulating DNA topology, are ubiquitous and indispensable for cellular survival due to the numerous roles they play during DNA metabolism. As we review here, current structural approaches have revealed unprecedented insights into the complex DNA-topoisomerase interaction and strand passage mechanism, helping to advance our understanding of their activities in vivo. This has been complemented by single-molecule techniques, which have facilitated the detailed dissection of the various topoisomerase reactions. Recent work has also revealed the importance of topoisomerase interactions with accessory proteins and other DNA-associated proteins, supporting the idea that they often function as part of multi-enzyme assemblies in vivo. In addition, novel topoisomerases have been identified and explored, such as topo VIII and Mini-A. These new findings are advancing our understanding of DNA-related processes and the vital functions topos fulfil, demonstrating their indispensability in virtually every aspect of DNA metabolism.
Keywords: DNA gyrase; DNA supercoiling; DNA topoisomerase; anti-cancer drugs; antibiotics.
© 2021 The Authors. BioEssays published by Wiley Periodicals LLC. This article has been contributed to by US Government employees and their work is in the public domain in the USA.
Conflict of interest statement
None declared.
Figures



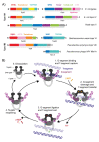


References
-
- Watson JD, Crick FH. Genetical implications of the structure of deoxyribonucleic acid. Nature. 1953;171(4361):964–967. - PubMed
-
- Watson JD, Crick FHC. A structure for deoxyribose nucleic acid. Nature. 1953;171:737–738. - PubMed
-
- Bates AD, Maxwell A. DNA Topology. Oxford University Press; Oxford: 2005.
Publication types
MeSH terms
Substances
Grants and funding
- 110072/Z/15/Z/WT_/Wellcome Trust/United Kingdom
- BB/P012523/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/J/00000201/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- ZIA HL001056/ImNIH/Intramural NIH HHS/United States
- 110072/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

