Identification of a Prognostic Colorectal Cancer Model Including LncRNA FOXP4-AS1 and LncRNA BBOX1-AS1 Based on Bioinformatics Analysis
- PMID: 33481661
- PMCID: PMC9805880
- DOI: 10.1089/cbr.2020.4242
Identification of a Prognostic Colorectal Cancer Model Including LncRNA FOXP4-AS1 and LncRNA BBOX1-AS1 Based on Bioinformatics Analysis
Abstract
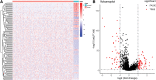
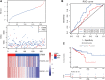
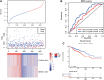
Background: Knowledge about the prognostic role of long noncoding RNA (lncRNA) in colorectal cancer (CRC) is limited. Therefore, we constructed a lncRNA-related prognostic model based on data from the Gene Expression Omnibus (GEO) and The Cancer Genome Atlas (TCGA). Materials and Methods: CRC transcriptome and clinical data were downloaded from the GSE20916 dataset and the TCGA database, respectively. R software was used for data processing and analysis. The differential lncRNA expression within the two datasets was first screened, and then intersections were measured. Cox regression and the Kaplan-Meier method were used to evaluate the effects of various factors on prognosis. The area under the curve (AUC) of the receiver operating characteristic curve and a nomogram based on multivariate Cox analysis were used to estimate the prognostic value of the lncRNA-related model. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were applied to elucidate the significantly involved biological functions and pathways. Results: A total of 11 lncRNAs were crossed. The univariate Cox analysis screened out two lncRNAs, which were analyzed in the multivariate Cox analysis. A nomogram based on the two lncRNAs and other clinicopathological risk factors was constructed. The AUC of the nomogram was 0.56 at 3 years and 0.71 at 5 years. The 3-year nomogram model was compared with the ideal model, which showed that some indices of the 3-year model were consistent with the ideal model, suggesting that our model was highly accurate. The GO and KEGG enrichment analyses showed that positive regulation of secretion by cells, positive regulation of secretion, positive regulation of exocytosis, endocytosis, and the calcium signaling pathway were differentially enriched in the two-lncRNA-associated phenotype. Conclusions: A two-lncRNA prognostic model of CRC was constructed by bioinformatics analysis. The model had moderate prediction accuracy. LncRNA BBOX1-AS1 and lncRNA FOXP4-AS1 were identified as prognostic biomarkers.
Keywords: bioinformatics analysis; colorectal cancer; lncRNA; prognostic model.
Conflict of interest statement
No competing financial interests exist.
Figures







Similar articles
-
Long non-coding RNA profile study identifies a metabolism-related signature for colorectal cancer.Mol Med. 2021 Aug 3;27(1):83. doi: 10.1186/s10020-021-00343-x. Mol Med. 2021. PMID: 34344319 Free PMC article.
-
Identification and Validation of Six Autophagy-related Long Non-coding RNAs as Prognostic Signature in Colorectal Cancer.Int J Med Sci. 2021 Jan 1;18(1):88-98. doi: 10.7150/ijms.49449. eCollection 2021. Int J Med Sci. 2021. PMID: 33390777 Free PMC article.
-
Identification of a five-long non-coding RNA signature to improve the prognosis prediction for patients with hepatocellular carcinoma.World J Gastroenterol. 2018 Aug 14;24(30):3426-3439. doi: 10.3748/wjg.v24.i30.3426. World J Gastroenterol. 2018. PMID: 30122881 Free PMC article.
-
Identification of MFI2-AS1, a Novel Pivotal lncRNA for Prognosis of Stage III/IV Colorectal Cancer.Dig Dis Sci. 2020 Dec;65(12):3538-3550. doi: 10.1007/s10620-020-06064-1. Epub 2020 Jan 20. Dig Dis Sci. 2020. PMID: 31960204 Review.
-
Prognostic effect of lncRNA BBOX1-AS1 in malignancies: a meta-analysis.Front Genet. 2023 Aug 11;14:1234040. doi: 10.3389/fgene.2023.1234040. eCollection 2023. Front Genet. 2023. PMID: 37636267 Free PMC article.
Cited by
-
Emerging roles of long non-coding RNA FOXP4-AS1 in human cancers: From molecular biology to clinical application.Heliyon. 2024 Oct 26;10(21):e39857. doi: 10.1016/j.heliyon.2024.e39857. eCollection 2024 Nov 15. Heliyon. 2024. PMID: 39539976 Free PMC article. Review.
-
FOXP4-AS1 May be a Potential Prognostic Biomarker in Human Cancers: A Meta-Analysis and Bioinformatics Analysis.Front Oncol. 2022 May 26;12:799265. doi: 10.3389/fonc.2022.799265. eCollection 2022. Front Oncol. 2022. PMID: 35719909 Free PMC article.
-
A review on the role of gamma-butyrobetaine hydroxylase 1 antisense RNA 1 in the carcinogenesis and tumor progression.Cancer Cell Int. 2023 Nov 5;23(1):263. doi: 10.1186/s12935-023-03113-3. Cancer Cell Int. 2023. PMID: 37925403 Free PMC article. Review.
-
LncRNA FOXP4-AS1 Promotes the Progression of Esophageal Squamous Cell Carcinoma by Interacting With MLL2/H3K4me3 to Upregulate FOXP4.Front Oncol. 2021 Dec 14;11:773864. doi: 10.3389/fonc.2021.773864. eCollection 2021. Front Oncol. 2021. PMID: 34970490 Free PMC article.
-
Activating transcription factor 3-activated long noncoding RNA forkhead box P4-antisense RNA 1 aggravates colorectal cancer progression by regulating microRNA-423-5p/nucleus accumbens associated 1 axis.Bioengineered. 2022 Feb;13(2):2114-2129. doi: 10.1080/21655979.2021.2023798. Bioengineered. 2022. PMID: 35034547 Free PMC article.
References
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7. - PubMed
-
- Siegel RL, Miller KD, Goding Sauer A, et al. . Colorectal cancer statistics, 2020. CA Cancer J Clin 2020;70:145. - PubMed
-
- Lee SJ, Lee J, Park SH, et al. . c-MET overexpression in colorectal cancer: A poor prognostic factor for survival. Clin Colorectal Cancer 2018;17:165. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
