Targeting TRIM Proteins: A Quest towards Drugging an Emerging Protein Class
- PMID: 33482040
- PMCID: PMC8251876
- DOI: 10.1002/cbic.202000787
Targeting TRIM Proteins: A Quest towards Drugging an Emerging Protein Class
Abstract
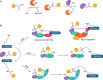
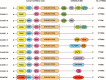
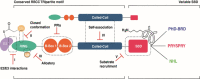
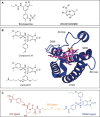
The ubiquitylation machinery regulates several fundamental biological processes from protein homeostasis to a wide variety of cellular signaling pathways. As a consequence, its dysregulation is linked to diseases including cancer, neurodegeneration, and autoimmunity. With this review, we aim to highlight the therapeutic potential of targeting E3 ligases, with a special focus on an emerging class of RING ligases, named tri-partite motif (TRIM) proteins, whose role as targets for drug development is currently gaining pharmaceutical attention. TRIM proteins exert their catalytic activity as scaffolds involved in many protein-protein interactions, whose multidomains and adapter-like nature make their druggability very challenging. Herein, we give an overview of the current understanding of this class of single polypeptide RING E3 ligases and discuss potential targeting options.
Keywords: high-throughput screening; inhibitors; ligases.
© 2021 The Authors. ChemBioChem published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

