Patterns of transcription factor programs and immune pathway activation define four major subtypes of SCLC with distinct therapeutic vulnerabilities
- PMID: 33482121
- PMCID: PMC8143037
- DOI: 10.1016/j.ccell.2020.12.014
Patterns of transcription factor programs and immune pathway activation define four major subtypes of SCLC with distinct therapeutic vulnerabilities
Abstract
Despite molecular and clinical heterogeneity, small cell lung cancer (SCLC) is treated as a single entity with predictably poor results. Using tumor expression data and non-negative matrix factorization, we identify four SCLC subtypes defined largely by differential expression of transcription factors ASCL1, NEUROD1, and POU2F3 or low expression of all three transcription factor signatures accompanied by an Inflamed gene signature (SCLC-A, N, P, and I, respectively). SCLC-I experiences the greatest benefit from the addition of immunotherapy to chemotherapy, while the other subtypes each have distinct vulnerabilities, including to inhibitors of PARP, Aurora kinases, or BCL-2. Cisplatin treatment of SCLC-A patient-derived xenografts induces intratumoral shifts toward SCLC-I, supporting subtype switching as a mechanism of acquired platinum resistance. We propose that matching baseline tumor subtype to therapy, as well as manipulating subtype switching on therapy, may enhance depth and duration of response for SCLC patients.
Keywords: ASCL1; EMT; NEUROD1; POU2F3; SCLC; intratumoral heterogeneity; neuroendocrine.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests C.M.G. reports research funding from AstraZeneca. S.H. reports consulting/honoraria from Qiagen, Boehringer Ingelheim and travel funding from Roche. B.Y.Z. and D.S.S. are employees of Genentech, Inc. G.F. is an employee of AstraZeneca and has stock ownership in AstraZeneca. B.G. reports research funding from ISA Pharm, Cue Bio, Pfizer, and Medimmune. I.I.W. reports consulting/advisory roles for Medscape, MSD, Genentech/Roche, PlatformQ Health, Pfizer, Bayer, Bristol-Myers Squibb, AstraZeneca/Medimmune, HTG Molecular, Asuragen, Merck, GlaxoSmithKline, Guardant Health, Oncocyte, and MSD, research support from Genentech, Oncoplex, HTG Molecular, DepArray, Merck, Bristol-Myers Squibb, Medimmune, Adapative, Adaptimmune, EMD Serono, Pfizer, Takeda, Amgen, Karus, Johnson & Johnson, Bayer, Iovance, 4D, Novartis and Akoya. J.V.H. serves on advisory committees for AstraZeneca, Bristol-Myers Squibb, GlaxoSmithKline, Guardant Health, Kairos Venture Investments, BrightPath Biotherapeutics, Hengrui Therapeutics, Eli Lilly, Spectrum, EMD Serono, Roche, and Foundation One Medicine and has research support from AstraZeneca, Spectrum, and Checkmate Pharmaceuticals, as well as royalties/licensing fees from Spectrum and Bio-Tree Systems. L.A.B. serves on advisory committees for AstraZeneca, AbbVie, GenMab, BergenBio, Pharma Mar SA, Sierra Oncology, Merck, Bristol-Myers Squibb, Genentech, and Pfizer and has research support from AbbVie, AstraZeneca, GenMab, Sierra Oncology, Tolero Pharmaceuticals. C.M.G., L.A.B., and J.V.H. have pending patent applications for SCLC subtyping U.S. 62/895,322. Otherwise, there are no pertinent financial or non-financial conflicts of interest to report.
Figures
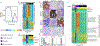






Comment in
-
Small cell lung cancer enters the era of precision medicine.Cancer Cell. 2021 Mar 8;39(3):297-299. doi: 10.1016/j.ccell.2021.02.002. Epub 2021 Feb 11. Cancer Cell. 2021. PMID: 33577787
References
-
- Allison Stewart C, Tong P, Cardnell RJ, Sen T, Li L, Gay CM, Masrorpour F, Fan Y, Bara RO, Feng Y, et al. (2017). Dynamic variations in epithelial-to-mesenchymal transition (EMT), ATM, and SLFN11 govern response to PARP inhibitors and cisplatin in small cell lung cancer. Oncotarget 8, 28575–28587. - PMC - PubMed
-
- Antonia SJ, Lopez-Martin JA, Bendell J, Ott PA, Taylor M, Eder JP, Jager D, Pietanza MC, Le DT, de Braud F, et al. (2016). Nivolumab alone and nivolumab plus ipilimumab in recurrent small-cell lung cancer (CheckMate 032): a multicentre, open-label, phase 1/2 trial. Lancet Oncol 17, 883–895. - PubMed
-
- Bonnafous CP, Valentine; Trichard Sylvia; Perrot Ivan; Cornen Stéphanie ; Morel Ariane; Breso Violette; Morel Yannis; Rossi Benjamin; Paturel Carine; Gauthier Laurent; Blery Mathieu (2013). Abstract 5037: Targeting MICA with therapeutic antibodies for the treatment of cancer. Journal for ImmunoTherapy of Cancer 1, P41.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

