The Genomic Landscape of Actinic Keratosis
- PMID: 33482222
- PMCID: PMC8221374
- DOI: 10.1016/j.jid.2020.12.024
The Genomic Landscape of Actinic Keratosis
Abstract
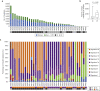
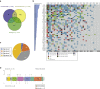
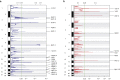
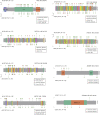
Actinic keratoses (AKs) are lesions of epidermal keratinocyte dysplasia and are precursors for invasive cutaneous squamous cell carcinoma (cSCC). Identifying the specific genomic alterations driving the progression from normal skin to skin with AK to skin with invasive cSCC is challenging because of the massive UVR-induced mutational burden characteristic at all stages of this progression. In this study, we report the largest AK whole-exome sequencing study to date and perform a mutational signature and candidate driver gene analysis on these lesions. We demonstrate in 37 AKs from both immunosuppressed and immunocompetent patients that there are significant similarities between AKs and cSCC in terms of mutational burden, copy number alterations, mutational signatures, and patterns of driver gene mutations. We identify 44 significantly mutated AK driver genes and confirm that these genes are similarly altered in cSCC. We identify azathioprine mutational signature in all AKs from patients exposed to the drug, providing further evidence for its role in keratinocyte carcinogenesis. cSCCs differ from AKs in having higher levels of intrasample heterogeneity. Alterations in signaling pathways also differ, with immune-related signaling and TGFβ signaling significantly more mutated in cSCC. Integrating our findings with independent gene expression datasets confirms that dysregulated TGFβ signaling may represent an important event in AK‒cSCC progression.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Figures





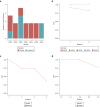
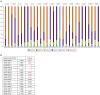
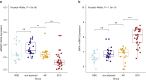

Comment in
-
Clarifying Progress on the Genomic Landscape of Actinic Keratosis.J Invest Dermatol. 2021 Jul;141(7):1622-1624. doi: 10.1016/j.jid.2021.02.761. J Invest Dermatol. 2021. PMID: 34167719
References
-
- Albibas A.A., Rose-Zerilli M.J.J., Lai C., Pengelly R.J., Lockett G.A., Theaker J. Subclonal evolution of cancer-related gene mutations in p53 immunopositive patches in human skin. J Invest Dermatol. 2018;138:189–198. - PubMed
-
- Ashton K.J., Weinstein S.R., Maguire D.J., Griffiths L.R. Chromosomal aberrations in squamous cell carcinoma and solar keratoses revealed by comparative genomic hybridization. Arch Dermatol. 2003;139:876–882. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

