Single-cell Immune Landscape of Human Recurrent Miscarriage
- PMID: 33482359
- PMCID: PMC8602400
- DOI: 10.1016/j.gpb.2020.11.002
Single-cell Immune Landscape of Human Recurrent Miscarriage
Abstract
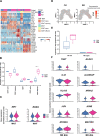
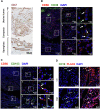
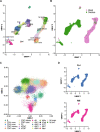
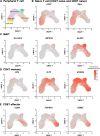
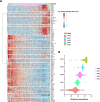
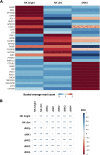
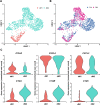
Successful pregnancy in placental mammals substantially depends on the establishment of maternal immune tolerance to the semi-allogenic fetus. Disorders in this process are tightly associated with adverse pregnancy outcomes including recurrent miscarriage (RM). However, an in-depth understanding of the systematic and decidual immune environment in RM remains largely lacking. In this study, we utilized single-cell RNA-sequencing (scRNA-seq) to comparably analyze the cellular and molecular signatures of decidual and peripheral leukocytes in normal and unexplained RM pregnancies at the early stage of gestation. Integrative analysis identifies 22 distinct cell clusters in total, and a dramatic difference in leukocyte subsets and molecular properties in RM cases is revealed. Specifically, the cytotoxic properties of CD8+ effector T cells, nature killer (NK), and mucosal-associated invariant T (MAIT) cells in peripheral blood indicates apparently enhanced pro-inflammatory status, and the population proportions and ligand-receptor interactions of the decidual leukocyte subsets demonstrate preferential immune activation in RM patients. The molecular features, spatial distribution, and the developmental trajectories of five decidual NK (dNK) subsets have been elaborately illustrated. In RM patients, a dNK subset that supports embryonic growth is diminished in proportion, while the ratio of another dNK subset with cytotoxic and immune-active signature is significantly increased. Notably, a unique pro-inflammatory CD56+CD16+ dNK subset substantially accumulates in RM decidua. These findings reveal a comprehensive cellular and molecular atlas of decidual and peripheral leukocytes in human early pregnancy and provide an in-depth insight into the immune pathogenesis for early pregnancy loss.
Keywords: Decidual and peripheral leukocytes; Developmental trajectory; Early pregnancy; Recurrent miscarriage; Single-cell RNA-sequencing.
Copyright © 2021 The Authors. Published by Elsevier B.V. All rights reserved.
Figures






References
-
- Erlebacher A. Immunology of the maternal-fetal interface. Annu Rev Immunol. 2013;31:387–411. - PubMed
-
- Kheshtchin N., Gharagozloo M., Andalib A., Ghahiri A., Maracy M.R., Rezaei A. The expression of Th1- and Th2-related chemokine receptors in women with recurrent miscarriage: the impact of lymphocyte immunotherapy. Am J Reprod Immunol. 2010;64:104–112. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

