Pharmacologic Normalization of Pancreatic Cancer-Associated Fibroblast Secretome Impairs Prometastatic Cross-Talk With Macrophages
- PMID: 33482394
- PMCID: PMC8024982
- DOI: 10.1016/j.jcmgh.2021.01.008
Pharmacologic Normalization of Pancreatic Cancer-Associated Fibroblast Secretome Impairs Prometastatic Cross-Talk With Macrophages
Abstract
Background & aims: Cancer-associated fibroblasts (CAFs) from pancreatic adenocarcinoma (PDA) present high protein synthesis rates. CAFs express the G-protein-coupled somatostatin receptor sst1. The sst1 agonist SOM230 blocks CAF protumoral features in vitro and in immunocompromised mice. We have explored here the therapeutic potential of SOM230, and underlying mechanisms, in immunocompetent models of murine PDA mimicking the heavy fibrotic and immunosuppressive stroma observed in patient tumors.
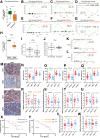
Methods: Large-scale mass spectrometry analyses were performed on media conditioned from 9 patient PDA-derived CAF primary cultures. Spontaneous transgenic and experimental (orthotopic co-graft of tumor cells plus CAFs) PDA-bearing mice were longitudinally ultrasound-monitored for tumor and metastatic progression. Histopathology and flow cytometry analyses were performed on primary tumors and metastases. Stromal signatures were functionally validated through bioinformatics using several published, and 1 original, PDA database.
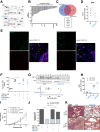
Results: Proteomics on the CAF secretome showed that SOM230 controls stromal activities including inflammatory responses. Among the identified secreted proteins, we validated that colony-stimulating factor 1 (CSF-1) (a macrophage growth factor) was reduced by SOM230 in the tumor and plasma of PDA-harboring mice, alongside intratumor stromal normalization (reduced CAF and macrophage activities), and dramatic metastasis reduction. In transgenic mice, these SOM230 benefits alleviate the chemotherapy-induced (gemcitabine) immunosuppressive stroma reshaping. Mechanistically, SOM230 acts in vivo on CAFs through sst1 to disrupt prometastatic CAF production of CSF-1 and cross-talk with macrophages. We found that in patients, stromal CSF-1 was associated with aggressive PDA forms.
Conclusions: We propose SOM230 as an antimetastatic therapy in PDA for its capacity to remodel the fibrotic and immunosuppressive myeloid stroma. This pharmacotherapy should benefit PDA patients treated with chemotherapies.
Keywords: Antimetastatic Therapy; Cancer-Associated Fibroblasts; Macrophages; Pancreatic Adenocarcinoma; Somatostatin Receptor; Stroma Normalization; Stromal Cell Cross-Talk.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Figures







References
-
- Kleeff J., Korc M., Apte M., La Vecchia C., Johnson C.D., Biankin A.V., Neale R.E., Tempero M., Tuveson D.A., Hruban R.H., Neoptolemos J.P. Pancreatic cancer. Nat Rev Dis Primers. 2016;2:16022. - PubMed
-
- Collisson E.A., Sadanandam A., Olson P., Gibb W.J., Truitt M., Gu S., Cooc J., Weinkle J., Kim G.E., Jakkula L., Feiler H.S., Ko A.H., Olshen A.B., Danenberg K.L., Tempero M.A., Spellman P.T., Hanahan D., Gray J.W. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med. 2011;17:500–503. - PMC - PubMed
-
- Bailey P., Chang D.K., Nones K., Johns A.L., Patch A.M., Gingras M.C., Miller D.K., Christ A.N., Bruxner T.J., Quinn M.C., Nourse C., Murtaugh L.C., Harliwong I., Idrisoglu S., Manning S., Nourbakhsh E., Wani S., Fink L., Holmes O., Chin V., Anderson M.J., Kazakoff S., Leonard C., Newell F., Waddell N., Wood S., Xu Q., Wilson P.J., Cloonan N., Kassahn K.S., Taylor D., Quek K., Robertson A., Pantano L., Mincarelli L., Sanchez L.N., Evers L., Wu J., Pinese M., Cowley M.J., Jones M.D., Colvin E.K., Nagrial A.M., Humphrey E.S., Chantrill L.A., Mawson A., Humphris J., Chou A., Pajic M., Scarlett C.J., Pinho A.V., Giry-Laterriere M., Rooman I., Samra J.S., Kench J.G., Lovell J.A., Merrett N.D., Toon C.W., Epari K., Nguyen N.Q., Barbour A., Zeps N., Moran-Jones K., Jamieson N.B., Graham J.S., Duthie F., Oien K., Hair J., Grutzmann R., Maitra A., Iacobuzio-Donahue C.A., Wolfgang C.L., Morgan R.A., Lawlor R.T., Corbo V., Bassi C., Rusev B., Capelli P., Salvia R., Tortora G., Mukhopadhyay D., Petersen G.M., Australian Pancreatic Cancer Genome I., Munzy D.M., Fisher W.E., Karim S.A., Eshleman J.R., Hruban R.H., Pilarsky C., Morton J.P., Sansom O.J., Scarpa A., Musgrove E.A., Bailey U.M., Hofmann O., Sutherland R.L., Wheeler D.A., Gill A.J., Gibbs R.A., Pearson J.V., Waddell N., Biankin A.V., Grimmond S.M. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47–52. - PubMed
-
- Moffitt R.A., Marayati R., Flate E.L., Volmar K.E., Loeza S.G., Hoadley K.A., Rashid N.U., Williams L.A., Eaton S.C., Chung A.H., Smyla J.K., Anderson J.M., Kim H.J., Bentrem D.J., Talamonti M.S., Iacobuzio-Donahue C.A., Hollingsworth M.A., Yeh J.J. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat Genet. 2015;47:1168–1178. - PMC - PubMed
-
- Nicolle R., Blum Y., Marisa L., Loncle C., Gayet O., Moutardier V., Turrini O., Giovannini M., Bian B., Bigonnet M., Rubis M., Elarouci N., Armenoult L., Ayadi M., Duconseil P., Gasmi M., Ouaissi M., Maignan A., Lomberk G., Boher J.M., Ewald J., Bories E., Garnier J., Goncalves A., Poizat F., Raoul J.L., Secq V., Garcia S., Grandval P., Barraud-Blanc M., Norguet E., Gilabert M., Delpero J.R., Roques J., Calvo E., Guillaumond F., Vasseur S., Urrutia R., de Reynies A., Dusetti N., Iovanna J. Pancreatic adenocarcinoma therapeutic targets revealed by tumor-stroma cross-talk analyses in patient-derived xenografts. Cell Rep. 2017;21:2458–2470. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

