Gene co-expression network analysis of Trypanosoma brucei in tsetse fly vector
- PMID: 33482903
- PMCID: PMC7821691
- DOI: 10.1186/s13071-021-04597-6
Gene co-expression network analysis of Trypanosoma brucei in tsetse fly vector
Abstract
Background: Trypanosoma brucei species are motile protozoan parasites that are cyclically transmitted by tsetse fly (genus Glossina) causing human sleeping sickness and nagana in livestock in sub-Saharan Africa. African trypanosomes display digenetic life cycle stages in the tsetse fly vector and in their mammalian host. Experimental work on insect-stage trypanosomes is challenging because of the difficulty in setting up successful in vitro cultures. Therefore, there is limited knowledge on the trypanosome biology during its development in the tsetse fly. Consequently, this limits the development of new strategies for blocking parasite transmission in the tsetse fly.
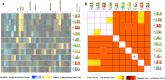
Methods: In this study, RNA-Seq data of insect-stage trypanosomes were used to construct a T. brucei gene co-expression network using the weighted gene co-expression analysis (WGCNA) method. The study identified significant enriched modules for genes that play key roles during the parasite's development in tsetse fly. Furthermore, potential 3' untranslated region (UTR) regulatory elements for genes that clustered in the same module were identified using the Finding Informative Regulatory Elements (FIRE) tool.
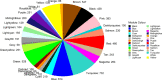
Results: A fraction of gene modules (12 out of 27 modules) in the constructed network were found to be enriched in functional roles associated with the cell division, protein biosynthesis, mitochondrion, and cell surface. Additionally, 12 hub genes encoding proteins such as RNA-binding protein 6 (RBP6), arginine kinase 1 (AK1), brucei alanine-rich protein (BARP), among others, were identified for the 12 significantly enriched gene modules. In addition, the potential regulatory elements located in the 3' untranslated regions of genes within the same module were predicted.
Conclusions: The constructed gene co-expression network provides a useful resource for network-based data mining to identify candidate genes for functional studies. This will enhance understanding of the molecular mechanisms that underlie important biological processes during parasite's development in tsetse fly. Ultimately, these findings will be key in the identification of potential molecular targets for disease control.
Keywords: Gene co-expression network; Trypanosoma brucei; Tsetse fly; Weighted gene co-expression network analysis.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Brun R, Schönenberger M. Cultivation and in vitro cloning or procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Short communication. Acta Trop. 1979;36:289–292. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

