Amoxicillin-Clavulanic Acid Resistance in the Genus Bifidobacterium
- PMID: 33483308
- PMCID: PMC8091617
- DOI: 10.1128/AEM.03137-20
Amoxicillin-Clavulanic Acid Resistance in the Genus Bifidobacterium
Abstract
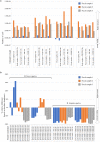
Amoxicillin-clavulanic acid (AMC) is one of the most frequently prescribed antibiotic formulations in the Western world. Extensive oral use of this antimicrobial combination influences the gut microbiota. One of the most abundant early colonizers of the human gut microbiota is represented by different taxa of the Bifidobacterium genus, which include many members that are considered to bestow beneficial effects upon their host. In the current study, we investigated the impact of AMC administration on the gut microbiota composition, comparing the gut microbiota of 23 children that had undergone AMC antibiotic therapy to that of 19 children that had not been treated with antibiotics during the preceding 6 months. Moreover, we evaluated AMC sensitivity by MIC test of 261 bifidobacterial strains, including reference strains for the currently recognized 64 bifidobacterial (sub)species, as well as 197 bifidobacterial isolates of human origin. These assessments allowed the identification of four bifidobacterial strains that exhibit a high level of AMC insensitivity, which were subjected to genomic and transcriptomic analyses to identify the putative genetic determinants responsible for this AMC insensitivity. Furthermore, we investigated the ecological role of AMC-resistant bifidobacterial strains by in vitro batch cultures.IMPORTANCE Based on our results, we observed a drastic reduction in gut microbiota diversity of children treated with antibiotics, which also affected the abundance of Bifidobacterium, a bacterial genus commonly found in the infant gut. MIC experiments revealed that more than 98% of bifidobacterial strains tested were shown to be inhibited by the AMC antibiotic. Isolation of four insensitive strains and sequencing of their genomes revealed the identity of possible genes involved in AMC resistance mechanisms. Moreover, gut-simulating in vitro experiments revealed that one strain, i.e., Bifidobacterium breve PRL2020, is able to persist in the presence of a complex microbiota combined with AMC antibiotic.
Keywords: antibiotics; bifidobacteria; comparative genomics.
Copyright © 2021 American Society for Microbiology.
Figures



References
-
- Turroni F, Peano C, Pass DA, Foroni E, Severgnini M, Claesson MJ, Kerr C, Hourihane J, Murray D, Fuligni F, Gueimonde M, Margolles A, De Bellis G, O’Toole PW, van Sinderen D, Marchesi JR, Ventura M. 2012. Diversity of bifidobacteria within the infant gut microbiota. PLoS One 7:e36957. doi:10.1371/journal.pone.0036957. - DOI - PMC - PubMed
-
- Milani C, Duranti S, Bottacini F, Casey E, Turroni F, Mahony J, Belzer C, Delgado Palacio S, Arboleya Montes S, Mancabelli L, Lugli GA, Rodriguez JM, Bode L, de Vos W, Gueimonde M, Margolles A, van Sinderen D, Ventura M. 2017. The first microbial colonizers of the human gut: composition, activities, and health implications of the infant gut microbiota. Microbiol Mol Biol Rev 81:e00036-17. doi:10.1128/MMBR.00036-17. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

