Deubiquitylating enzymes in neuronal health and disease
- PMID: 33483467
- PMCID: PMC7822931
- DOI: 10.1038/s41419-020-03361-5
Deubiquitylating enzymes in neuronal health and disease
Abstract
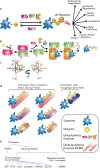
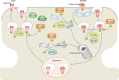
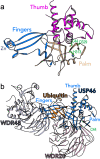
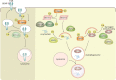
Ubiquitylation and deubiquitylation play a pivotal role in protein homeostasis (proteostasis). Proteostasis shapes the proteome landscape in the human brain and its impairment is linked to neurodevelopmental and neurodegenerative disorders. Here we discuss the emerging roles of deubiquitylating enzymes in neuronal function and survival. We provide an updated perspective on the genetics, physiology, structure, and function of deubiquitylases in neuronal health and disease.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

Similar articles
-
The pivotal role of ubiquitin-activating enzyme E1 (UBA1) in neuronal health and neurodegeneration.Int J Biochem Cell Biol. 2020 Jun;123:105746. doi: 10.1016/j.biocel.2020.105746. Epub 2020 Apr 18. Int J Biochem Cell Biol. 2020. PMID: 32315770 Review.
-
Distinct regulatory ribosomal ubiquitylation events are reversible and hierarchically organized.Elife. 2020 Feb 3;9:e54023. doi: 10.7554/eLife.54023. Elife. 2020. PMID: 32011234 Free PMC article.
-
A portrayal of E3 ubiquitin ligases and deubiquitylases in cancer.Int J Cancer. 2013 Dec 15;133(12):2759-68. doi: 10.1002/ijc.28129. Epub 2013 Mar 25. Int J Cancer. 2013. PMID: 23436247 Review.
-
Progressing neurobiological strategies against proteostasis failure: Challenges in neurodegeneration.Prog Neurobiol. 2017 Dec;159:1-38. doi: 10.1016/j.pneurobio.2017.08.005. Epub 2017 Sep 1. Prog Neurobiol. 2017. PMID: 28870769 Review.
-
La FAM fatale: USP9X in development and disease.Cell Mol Life Sci. 2015 Jun;72(11):2075-89. doi: 10.1007/s00018-015-1851-0. Epub 2015 Feb 12. Cell Mol Life Sci. 2015. PMID: 25672900 Free PMC article. Review.
Cited by
-
Enzymatically catalyzed molecular aggregation.Nat Commun. 2024 Nov 19;15(1):9999. doi: 10.1038/s41467-024-54291-1. Nat Commun. 2024. PMID: 39557870 Free PMC article.
-
Co-expression-wide association studies link genetically regulated interactions with complex traits.medRxiv [Preprint]. 2025 Jun 23:2024.10.02.24314813. doi: 10.1101/2024.10.02.24314813. medRxiv. 2025. PMID: 39711708 Free PMC article. Preprint.
-
Deubiquitinases as novel therapeutic targets for diseases.MedComm (2020). 2024 Dec 13;5(12):e70036. doi: 10.1002/mco2.70036. eCollection 2024 Dec. MedComm (2020). 2024. PMID: 39678489 Free PMC article. Review.
-
Ubiquitin Carboxyl-Terminal Hydrolase L1 and Its Role in Parkinson's Disease.Int J Mol Sci. 2024 Jan 21;25(2):1303. doi: 10.3390/ijms25021303. Int J Mol Sci. 2024. PMID: 38279302 Free PMC article. Review.
-
On the Study of Deubiquitinases: Using the Right Tools for the Job.Biomolecules. 2022 May 14;12(5):703. doi: 10.3390/biom12050703. Biomolecules. 2022. PMID: 35625630 Free PMC article. Review.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

