CRISPR technologies and the search for the PAM-free nuclease
- PMID: 33483498
- PMCID: PMC7822910
- DOI: 10.1038/s41467-020-20633-y
CRISPR technologies and the search for the PAM-free nuclease
Abstract
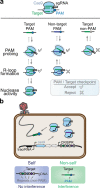
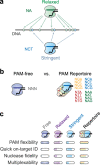
The ever-expanding set of CRISPR technologies and their programmable RNA-guided nucleases exhibit remarkable flexibility in DNA targeting. However, this flexibility comes with an ever-present constraint: the requirement for a protospacer adjacent motif (PAM) flanking each target. While PAMs play an essential role in self/nonself discrimination by CRISPR-Cas immune systems, this constraint has launched a far-reaching expedition for nucleases with relaxed PAM requirements. Here, we review ongoing efforts toward realizing PAM-free nucleases through natural ortholog mining and protein engineering. We also address potential consequences of fully eliminating PAM recognition and instead propose an alternative nuclease repertoire covering all possible PAM sequences.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

