The Drosophila melanogaster Neprilysin Nepl15 is involved in lipid and carbohydrate storage
- PMID: 33483521
- PMCID: PMC7822871
- DOI: 10.1038/s41598-021-81165-z
The Drosophila melanogaster Neprilysin Nepl15 is involved in lipid and carbohydrate storage
Erratum in
-
Author Correction: The Drosophila melanogaster Neprilysin Nepl15 is involved in lipid and carbohydrate storage.Sci Rep. 2021 Feb 25;11(1):5093. doi: 10.1038/s41598-021-84100-4. Sci Rep. 2021. PMID: 33633243 Free PMC article. No abstract available.
Abstract
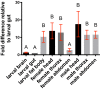
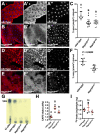
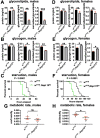
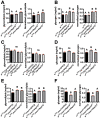
The prototypical M13 peptidase, human Neprilysin, functions as a transmembrane "ectoenzyme" that cleaves neuropeptides that regulate e.g. glucose metabolism, and has been linked to type 2 diabetes. The M13 family has undergone a remarkable, and conserved, expansion in the Drosophila genus. Here, we describe the function of Drosophila melanogaster Neprilysin-like 15 (Nepl15). Nepl15 is likely to be a secreted protein, rather than a transmembrane protein. Nepl15 has changes in critical catalytic residues that are conserved across the Drosophila genus and likely renders the Nepl15 protein catalytically inactive. Nevertheless, a knockout of the Nepl15 gene reveals a reduction in triglyceride and glycogen storage, with the effects likely occurring during the larval feeding period. Conversely, flies overexpressing Nepl15 store more triglycerides and glycogen. Protein modeling suggests that Nepl15 is able to bind and sequester peptide targets of catalytically active Drosophila M13 family members, peptides that are conserved in humans and Drosophila, potentially providing a novel mechanism for regulating the activity of neuropeptides in the context of lipid and carbohydrate homeostasis.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

