Mechanobiological conditioning of mesenchymal stem cells for enhanced vascular regeneration
- PMID: 33483713
- PMCID: PMC8875880
- DOI: 10.1038/s41551-020-00674-w
Mechanobiological conditioning of mesenchymal stem cells for enhanced vascular regeneration
Abstract
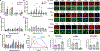
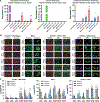
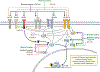
Using endogenous mesenchymal stem cells for treating myocardial infarction and other cardiovascular conditions typically results in poor efficacy, in part owing to the heterogeneity of the harvested cells and of the patient responses. Here, by means of high-throughput screening of the combinatorial space of mechanical-strain level and of the presence of particular kinase inhibitors, we show that human mesenchymal stem cells can be mechanically and pharmacologically conditioned to enhance vascular regeneration in vivo. Mesenchymal stem cells conditioned to increase the activation of signalling pathways mediated by Smad2/3 (mothers against decapentaplegic homolog 2/3) and YAP (Yes-associated protein) expressed markers that are associated with pericytes and endothelial cells, displayed increased angiogenic activity in vitro, and enhanced the formation of vasculature in mice after subcutaneous implantation and after implantation in ischaemic hindlimbs. These effects were mediated by the crosstalk of endothelial-growth-factor receptors, transforming-growth-factor-beta receptor type 1 and vascular-endothelial-growth-factor receptor 2. Mechanical and pharmacological conditioning can significantly enhance the regenerative properties of mesenchymal stem cells.
Figures



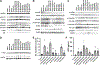
Comment in
-
Stretch-boosted cell-mediated vascularization.Nat Biomed Eng. 2021 Jan;5(1):6-7. doi: 10.1038/s41551-020-00680-y. Nat Biomed Eng. 2021. PMID: 33483711 No abstract available.
-
Boosting stem cell vascular regenerative capacity.Nat Rev Cardiol. 2021 May;18(5):306. doi: 10.1038/s41569-021-00525-4. Nat Rev Cardiol. 2021. PMID: 33542521 No abstract available.
References
-
- Mao Q, Liang XL, Wu YF, Pang YH, Zhao XJ and Lu YX. ILK promotes survival and self-renewal of hypoxic MSCs via the activation of lncTCF7-Wnt pathway induced by IL6/STAT3 signaling. Gene Ther. 2019;26:165–176. - PubMed
-
- Shi Y, Hu G, Su J, Li W, Chen Q, Shou P, Xu C, Chen X, Huang Y, Zhu Z, Huang X, Han X, Xie N and Ren G. Mesenchymal stem cells: a new strategy for immunosuppression and tissue repair. Cell Res. 2010;20:510–8. - PubMed
-
- Shake JG, Gruber PJ, Baumgartner WA, Senechal G, Meyers J, Redmond JM, Pittenger MF and Martin BJ. Mesenchymal stem cell implantation in a swine myocardial infarct model: engraftment and functional effects. Ann Thorac Surg. 2002;73:1919–25; discussion 1926. - PubMed
-
- Barbash IM, Chouraqui P, Baron J, Feinberg MS, Etzion S, Tessone A, Miller L, Guetta E, Zipori D, Kedes LH, Kloner RA and Leor J. Systemic delivery of bone marrow-derived mesenchymal stem cells to the infarcted myocardium: feasibility, cell migration, and body distribution. Circulation. 2003;108:863–8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

