Building pan-genome infrastructures for crop plants and their use in association genetics
- PMID: 33484244
- PMCID: PMC7934568
- DOI: 10.1093/dnares/dsaa030
Building pan-genome infrastructures for crop plants and their use in association genetics
Abstract
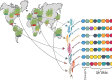
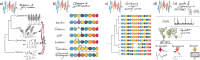
Pan-genomic studies aim at representing the entire sequence diversity within a species to provide useful resources for evolutionary studies, functional genomics and breeding of cultivated plants. Cost reductions in high-throughput sequencing and advances in sequence assembly algorithms have made it possible to create multiple reference genomes along with a catalogue of all forms of genetic variations in plant species with large and complex or polyploid genomes. In this review, we summarize the current approaches to building pan-genomes as an in silico representation of plant sequence diversity and outline relevant methods for their effective utilization in linking structural with phenotypic variation. We propose as future research avenues (i) transcriptomic and epigenomic studies across multiple reference genomes and (ii) the development of user-friendly and feature-rich pan-genome browsers.
Keywords: association genetics; crop plants; genome sequencing; genomics; pan-genome.
© The Author(s) 2021. Published by Oxford University Press on behalf of Kazusa DNA Research Institute.
Figures

References
-
- Esquinas-Alcázar J. 2005, Science and society: protecting crop genetic diversity for food security: political, ethical and technical challenges, Nat. Rev. Genet., 6, 946–53. - PubMed
-
- Dempewolf H., Bordoni P., Rieseberg L.H., et al.2010, Food security: crop species diversity, Science, 328, 169–70. - PubMed
-
- Godfray H.C.J., Beddington J.R., Crute I.R., et al.2010, Food security: the challenge of feeding 9 billion people, Science, 327, 812–8. - PubMed
-
- Mérot C., Oomen R.A., Tigano A., et al.2020, A roadmap for understanding the evolutionary significance of structural genomic variation, Trends Ecol. Evol., 35, 561–72. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

