microRNA-31 regulates skeletogenesis by direct suppression of Eve and Wnt1
- PMID: 33484703
- PMCID: PMC7956219
- DOI: 10.1016/j.ydbio.2021.01.008
microRNA-31 regulates skeletogenesis by direct suppression of Eve and Wnt1
Abstract
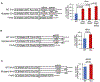
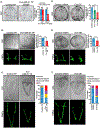
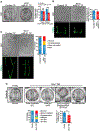
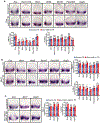
microRNAs (miRNAs) play a critical role in a variety of biological processes, including embryogenesis and the physiological functions of cells. Evolutionarily conserved microRNA-31 (miR-31) has been found to be involved in cancer, bone formation, and lymphatic development. We previously discovered that, in the sea urchin, miR-31 knockdown (KD) embryos have shortened dorsoventral connecting rods, mispatterned skeletogenic primary mesenchyme cells (PMCs) and shifted and expanded Vegf3 expression domain. Vegf3 itself does not contain miR-31 binding sites; however, we identified its upstream regulators Eve and Wnt1 to be directly suppressed by miR-31. Removal of miR-31's suppression of Eve and Wnt1 resulted in skeletal and PMC patterning defects, similar to miR-31 KD phenotypes. Additionally, removal of miR-31's suppression of Eve and Wnt1 results in an expansion and anterior shift in expression of Veg1 ectodermal genes, including Vegf3 in the blastulae. This indicates that miR-31 indirectly regulates Vegf3 expression through directly suppressing Eve and Wnt1. Furthermore, removing miR-31 suppression of Eve is sufficient to cause skeletogenic defects, revealing a novel regulatory role of Eve in skeletogenesis and PMC patterning. Overall, this study provides a proposed molecular mechanism of miR-31's regulation of skeletogenesis and PMC patterning through its cross-regulation of a Wnt signaling ligand and a transcription factor of the endodermal and ectodermal gene regulatory network.
Keywords: Even-skipped; MicroRNA-31; Post-transcriptional regulation; Primary mesenchyme cells; Sea urchin; Vegf signaling; Wnt; miRNA target protector.
Copyright © 2021 Elsevier Inc. All rights reserved.
Figures





Similar articles
-
microRNA-31 modulates skeletal patterning in the sea urchin embryo.Development. 2015 Nov 1;142(21):3769-80. doi: 10.1242/dev.127969. Epub 2015 Sep 23. Development. 2015. PMID: 26400092 Free PMC article.
-
microRNA-1 regulates sea urchin skeletogenesis by directly targeting skeletogenic genes and modulating components of signaling pathways.Dev Biol. 2024 Apr;508:123-137. doi: 10.1016/j.ydbio.2024.01.010. Epub 2024 Jan 28. Dev Biol. 2024. PMID: 38290645 Free PMC article.
-
Global analysis of primary mesenchyme cell cis-regulatory modules by chromatin accessibility profiling.BMC Genomics. 2018 Mar 20;19(1):206. doi: 10.1186/s12864-018-4542-z. BMC Genomics. 2018. PMID: 29558892 Free PMC article.
-
Embryonic pattern formation without morphogens.Bioessays. 2008 May;30(5):412-7. doi: 10.1002/bies.20758. Bioessays. 2008. PMID: 18404688 Review.
-
Gene regulatory networks for development.Proc Natl Acad Sci U S A. 2005 Apr 5;102(14):4936-42. doi: 10.1073/pnas.0408031102. Epub 2005 Mar 23. Proc Natl Acad Sci U S A. 2005. PMID: 15788537 Free PMC article. Review.
Cited by
-
Comparative Analysis of Bivalve and Sea Urchin Genetics and Development: Investigating the Dichotomy in Bilateria.Int J Mol Sci. 2023 Dec 5;24(24):17163. doi: 10.3390/ijms242417163. Int J Mol Sci. 2023. PMID: 38138992 Free PMC article. Review.
-
miR-31-mediated local translation at the mitotic spindle is important for early development.Development. 2024 Sep 1;151(17):dev202619. doi: 10.1242/dev.202619. Epub 2024 Sep 5. Development. 2024. PMID: 39250531 Free PMC article.
-
Distinct regulatory states control the elongation of individual skeletal rods in the sea urchin embryo.Dev Dyn. 2022 Aug;251(8):1322-1339. doi: 10.1002/dvdy.474. Epub 2022 Apr 22. Dev Dyn. 2022. PMID: 35403290 Free PMC article.
-
Effect of Anti-Osteoporotic Treatments on Circulating and Bone MicroRNA Patterns in Osteopenic ZDF Rats.Int J Mol Sci. 2022 Jun 10;23(12):6534. doi: 10.3390/ijms23126534. Int J Mol Sci. 2022. PMID: 35742976 Free PMC article.
-
microRNA-124 directly suppresses Nodal and Notch to regulate mesodermal development.Dev Biol. 2023 Oct;502:50-62. doi: 10.1016/j.ydbio.2023.06.017. Epub 2023 Jul 5. Dev Biol. 2023. PMID: 37419400 Free PMC article.
References
-
- Adomako-Ankomah A, Ettensohn CA, 2011. P58-A and P58-B: novel proteins that mediate skeletogenesis in the sea urchin embryo. Dev Biol 353, 81–93. - PubMed
-
- Adomako-Ankomah A, Ettensohn CA, 2013. Growth factor-mediated mesodermal cell guidance and skeletogenesis during sea urchin gastrulation. Development 140, 4214–4225. - PubMed
-
- Adomako-Ankomah A, Ettensohn CA, 2014. Growth factors and early mesoderm morphogenesis: insights from the sea urchin embryo. Genesis 52, 158–172. - PubMed
-
- Anstrom JA, 1992. Microfilaments, cell shape changes, and the formation of primary mesenchyme in sea urchin embryos. J Exp Zool 264, 312–322. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

