Small-molecule control of neurotransmitter sulfonation
- PMID: 33485192
- PMCID: PMC7948405
- DOI: 10.1074/jbc.RA120.015177
Small-molecule control of neurotransmitter sulfonation
Abstract
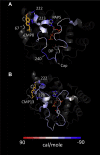
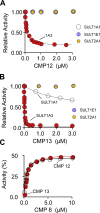
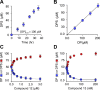
Controlling unmodified serotonin levels in brain synapses is a primary objective when treating major depressive disorder-a disease that afflicts ∼20% of the world's population. Roughly 60% of patients respond poorly to first-line treatments and thus new therapeutic strategies are sought. To this end, we have constructed isoform-specific inhibitors of the human cytosolic sulfotransferase 1A3 (SULT1A3)-the isoform responsible for sulfonating ∼80% of the serotonin in the extracellular brain fluid. The inhibitor design includes a core ring structure, which anchors the inhibitor into a SULT1A3-specific binding pocket located outside the active site, and a side chain crafted to act as a latch to inhibit turnover by fastening down the SULT1A3 active-site cap. The inhibitors are allosteric, they bind with nanomolar affinity and are highly specific for the 1A3 isoform. The cap-stabilizing effects of the latch can be accurately calculated and are predicted to extend throughout the cap and into the surrounding protein. A free-energy correlation demonstrates that the percent inhibition at saturating inhibitor varies linearly with cap stabilization - the correlation is linear because the rate-limiting step of the catalytic cycle, nucleotide release, scales linearly with the fraction of enzyme in the cap-open form. Inhibitor efficacy in cultured cells was studied using a human mammary epithelial cell line that expresses SULT1A3 at levels comparable with those found in neurons. The inhibitors perform similarly in ex vivo and in vitro studies; consequently, SULT1A3 turnover can now be potently suppressed in an isoform-specific manner in human cells.
Keywords: SULT1A3; allosteric; catecholamine; human mammary epithelial cells; inhibitor; molecular dynamics; neurotransmitter; structure activity relationship; sulfotransferase.
Copyright © 2020 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest A provisional patent covering the compounds described in this manuscript has been submitted (US patent application number PCT/US2019/065442). All authors listed on the masthead are co-inventors on the patent.
Figures



References
-
- Merikangas K.R., He J.P., Burstein M., Swanson S.A., Avenevoli S., Cui L., Benjet C., Georgiades K., Swendsen J. Lifetime prevalence of mental disorders in U.S. adolescents: results from the National Comorbidity Survey Replication--Adolescent Supplement (NCS-A) J. Am. Acad. Child. Adolesc. Psychiatry. 2010;49:980–989. - PMC - PubMed
-
- Souery D., Amsterdam J., de Montigny C., Lecrubier Y., Montgomery S., Lipp O., Racagni G., Zohar J., Mendlewicz J. Treatment resistant depression: methodological overview and operational criteria. Eur. Neuropsychopharmacol. 1999;9:83–91. - PubMed
-
- Thomas S.J., Shin M., McInnis M.G., Bostwick J.R. Combination therapy with monoamine oxidase inhibitors and other antidepressants or stimulants: strategies for the management of treatment-resistant depression. Pharmacotherapy. 2015;35:433–449. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

