Comprehensive metabolic profiling of Parkinson's disease by liquid chromatography-mass spectrometry
- PMID: 33485385
- PMCID: PMC7825156
- DOI: 10.1186/s13024-021-00425-8
Comprehensive metabolic profiling of Parkinson's disease by liquid chromatography-mass spectrometry
Abstract
Background: Parkinson's disease (PD) is a prevalent neurological disease in the elderly with increasing morbidity and mortality. Despite enormous efforts, rapid and accurate diagnosis of PD is still compromised. Metabolomics defines the final readout of genome-environment interactions through the analysis of the entire metabolic profile in biological matrices. Recently, unbiased metabolic profiling of human sample has been initiated to identify novel PD metabolic biomarkers and dysfunctional metabolic pathways, however, it remains a challenge to define reliable biomarker(s) for clinical use.
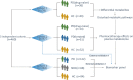
Methods: We presented a comprehensive metabolic evaluation for identifying crucial metabolic disturbances in PD using liquid chromatography-high resolution mass spectrometry-based metabolomics approach. Plasma samples from 3 independent cohorts (n = 460, 223 PD, 169 healthy controls (HCs) and 68 PD-unrelated neurological disease controls) were collected for the characterization of metabolic changes resulted from PD, antiparkinsonian treatment and potential interferences of other diseases. Unbiased multivariate and univariate analyses were performed to determine the most promising metabolic signatures from all metabolomic datasets. Multiple linear regressions were applied to investigate the associations of metabolites with age, duration time and stage of PD. The combinational biomarker model established by binary logistic regression analysis was validated by 3 cohorts.
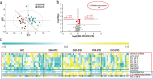
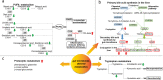
Results: A list of metabolites including amino acids, acylcarnitines, organic acids, steroids, amides, and lipids from human plasma of 3 cohorts were identified. Compared with HC, we observed significant reductions of fatty acids (FFAs) and caffeine metabolites, elevations of bile acids and microbiota-derived deleterious metabolites, and alterations in steroid hormones in drug-naïve PD. Additionally, we found that L-dopa treatment could affect plasma metabolome involved in phenylalanine and tyrosine metabolism and alleviate the elevations of bile acids in PD. Finally, a metabolite panel of 4 biomarker candidates, including FFA 10:0, FFA 12:0, indolelactic acid and phenylacetyl-glutamine was identified based on comprehensive discovery and validation workflow. This panel showed favorable discriminating power for PD.
Conclusions: This study may help improve our understanding of PD etiopathogenesis and facilitate target screening for therapeutic intervention. The metabolite panel identified in this study may provide novel approach for the clinical diagnosis of PD in the future.
Keywords: Bile acid profile; Biomarker; Metabolic disturbances; Metabolomics; Parkinson’s disease.
Conflict of interest statement
The author declares no competing interests.
Figures



References
-
- Cressatti M, Juwara L, Galindez JM, Velly AM, Nkurunziza ES, Marier S, Canie O, Gornistky M, Schipper HM. Salivary microR-153 and microR-223 levels as potential diagnostic biomarkers of idiopathic Parkinson's disease. Mov Disord. 2020;35:468-77. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

