Apolipoprotein E Polymorphism and Oxidative Stress in Human Peripheral Blood Cells: Can Physical Activity Reactivate the Proteasome System through Epigenetic Mechanisms?
- PMID: 33488947
- PMCID: PMC7796851
- DOI: 10.1155/2021/8869849
Apolipoprotein E Polymorphism and Oxidative Stress in Human Peripheral Blood Cells: Can Physical Activity Reactivate the Proteasome System through Epigenetic Mechanisms?
Abstract
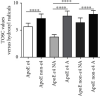
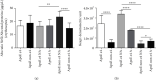
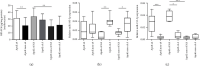
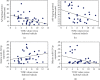
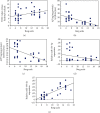
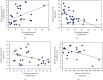
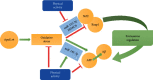
Alzheimer's disease (AD) is characterized by proteasome activity impairment, oxidative stress, and epigenetic changes, resulting in β-amyloid (Aβ) production/degradation imbalance. Apolipoprotein E (ApoE) is implicated in Aβ clearance, and particularly, the ApoE ε4 isoform predisposes to AD development. Regular physical activity is known to reduce AD progression. However, the impact of ApoE polymorphism and physical exercise on Aβ production and proteasome system activity has never been investigated in human peripheral blood cells, particularly in erythrocytes, an emerging peripheral model used to study biochemical alteration. Therefore, the influence of ApoE polymorphism on the antioxidant defences, amyloid accumulation, and proteasome activity was here evaluated in human peripheral blood cells depending on physical activity, to assess putative peripheral biomarkers for AD and candidate targets that could be modulated by lifestyle. Healthy subjects were enrolled and classified based on the ApoE polymorphism (by the restriction fragment length polymorphism technique) and physical activity level (Borg scale) and grouped into ApoE ε4/non-ε4 carriers and active/non-active subjects. The plasma antioxidant capability (AOC), the erythrocyte Aβ production/accumulation, and the nuclear factor erythroid 2-related factor 2 (Nrf2) mediated proteasome functionality were evaluated in all groups by the chromatographic and immunoenzymatic assay, respectively. Moreover, epigenetic mechanisms were investigated considering the expression of histone deacetylase 6, employing a competitive ELISA, and the modulation of two key miRNAs (miR-153-3p and miR-195-5p), through the miRNeasy Serum/Plasma Mini Kit. ApoE ε4 subjects showed a reduction in plasma AOC and an increase in the Nrf2 blocker, miR-153-3p, contributing to an enhancement of the erythrocyte concentration of Aβ. Physical exercise increased plasma AOC and reduced the amount of Aβ and its precursor, involving a reduced miR-153-3p expression and a miR-195-5p enhancement. Our data highlight the impact of the ApoE genotype on the amyloidogenic pathway and the proteasome system, suggesting the positive impact of physical exercise, also through epigenetic mechanisms.
Copyright © 2021 Rebecca Piccarducci et al.
Conflict of interest statement
The authors declare that there is no conflict of interest regarding the publication of this paper.
Figures
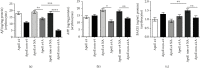
References
-
- Baars H. F., Doevendans P. A. F. M., van der Smagt J. J. Clinical Cardiogenetics. London: Springer-Verlag; 2011. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

