Studying Histone Deacetylase Inhibition and Apoptosis Induction of Psammaplin A Monomers with Modified Thiol Group
- PMID: 33488962
- PMCID: PMC7812609
- DOI: 10.1021/acsmedchemlett.0c00369
Studying Histone Deacetylase Inhibition and Apoptosis Induction of Psammaplin A Monomers with Modified Thiol Group
Abstract
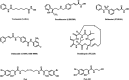
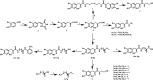
Psammaplin A (PsA) is a bromotyrosine disulfide dimer with histone deacetylase (HDAC) inhibition and acts through reduced monomer PsA-SH. We studied the connection of HDAC inhibition, cell growth inhibition, and apoptosis induction of PsA-SH by modifying the -SH group with deletion (6a) or replacement with hydroxamic acid (10b) or benzamide (12g). PsA-SH inhibits HDAC1/2/3 and 6a loses the HDAC inhibition ability. 10b inhibits HDAC1/2/3/6 while 12g shows selective inhibition of HDAC3. PsA-SH and 10b, but neither 6a nor 12g, induce apoptosis in human leukemia HL-60 cells associated with increased acetylation of Histone H3. PsA-SH and 10b inhibit growth of several solid tumor cell lines in vitro and Lewis lung cancer cell growth in vivo. PsA-SH is a simple scaffold for developing selective HDAC inhibitors and induces apoptosis through inhibiting HDAC1/2.
© 2021 American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures





References
-
- Garmpis N.; Damaskos C.; Garmpi A.; Dimitroulis D.; Spartalis E.; Margonis G. A.; Schizas D.; Deskou I.; Doula C.; Magkouti E.; Andreatos N.; Antoniou E. A.; Nonni A.; Kontzoglou K.; Mantas D. Targeting Histone Deacetylases in Malignant Melanoma: A Future Therapeutic Agent or Just Great Expectations?. Anticancer research 2017, 37, 5355–5362. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information
Research Materials
Miscellaneous

