Solution Conformations Shed Light on PROTAC Cell Permeability
- PMID: 33488971
- PMCID: PMC7812666
- DOI: 10.1021/acsmedchemlett.0c00556
Solution Conformations Shed Light on PROTAC Cell Permeability
Abstract
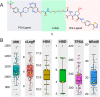
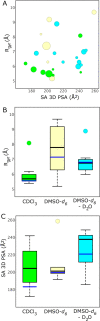
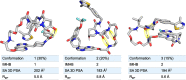
Proteolysis targeting chimeras (PROTACs) induce intracellular degradation of target proteins. Their bifunctional structure puts degraders in a chemical space where ADME properties often complicate drug discovery. Herein we provide the first structural insight into PROTAC cell permeability obtained by NMR studies of a VHL-based PROTAC (1), which is cell permeable despite having a high molecular weight and polarity and a large number of rotatable bonds. We found that 1 populates elongated and polar conformations in solutions that mimic extra- and intracellular compartments. Conformations were folded and had a smaller polar surface area in chloroform, mimicking a cell membrane interior. Formation of intramolecular and nonclassical hydrogen bonds, π-π interactions, and shielding of amide groups from solvent all facilitate cell permeability by minimization of size and polarity. We conclude that molecular chameleonicity appears to be of major importance for 1 to enter into target cells.
© 2020 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare the following competing financial interest(s): D.G. is an employee of Nuvisan Innovation Campus Berlin GmbH. A.G and D.M. are employees of Bayer AG.
Figures



References
-
- Bondeson D. P.; Mares A.; Smith I. E. D.; Ko E.; Campos S.; Miah A. H.; Mulholland K. E.; Routly N.; Buckley D. L.; Gustafson J. L.; Zinn N.; Grandi P.; Shimamura S.; Bergamini G.; Faelth-Savitski M.; Bantscheff M.; Cox C.; Gordon D. A.; Willard R. R.; Flanagan J. J.; Casillas L. N.; Votta B. J.; den Besten W.; Famm K.; Kruidenier L.; Carter P. S.; Harling J. D.; Churcher I.; Crews C. M. Catalytic in vivo protein knockdown by small-molecule PROTACs. Nat. Chem. Biol. 2015, 11, 611–617. 10.1038/nchembio.1858. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information
Research Materials
Miscellaneous

