Investigating Major Recurring Campylobacter jejuni Lineages in Luxembourg Using Four Core or Whole Genome Sequencing Typing Schemes
- PMID: 33489938
- PMCID: PMC7819963
- DOI: 10.3389/fcimb.2020.608020
Investigating Major Recurring Campylobacter jejuni Lineages in Luxembourg Using Four Core or Whole Genome Sequencing Typing Schemes
Abstract
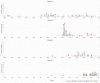
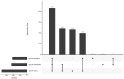
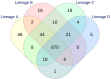
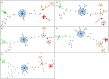
Campylobacter jejuni is the leading cause of bacterial gastroenteritis, which has motivated the monitoring of genetic profiles circulating in Luxembourg since 13 years. From our integrated surveillance using a genotyping strategy based on an extended MLST scheme including gyrA and porA markers, an unexpected endemic pattern was discovered in the temporal distribution of genotypes. We aimed to test the hypothesis of stable lineages occurrence by implementing whole genome sequencing (WGS) associated with comprehensive and internationally validated schemes. This pilot study assessed four WGS-based typing schemes to classify a panel of 108 strains previously identified as recurrent or sporadic profiles using this in-house typing system. The strain collection included four common lineages in human infection (N = 67) initially identified from recurrent combination of ST-gyrA-porA alleles also detected in non-human samples: veterinary (N = 19), food (N = 20), and environmental (N = 2) sources. An additional set of 19 strains belonging to sporadic profiles completed the tested panel. All the strains were processed by WGS by using Illumina technologies and by applying stringent criteria for filtering sequencing data; we ensure robustness in our genomic comparison. Four typing schemes were applied to classify the strains: (i) the cgMLST SeqSphere+ scheme of 637 loci, (ii) the cgMLST Oxford scheme of 1,343 loci, (iii) the cgMLST INNUENDO scheme of 678 loci, and (iv) the wgMLST INNUENDO scheme of 2,795 loci. A high concordance between the typing schemes was determined by comparing the calculated adjusted Wallace coefficients. After quality control and analyses with these four typing schemes, 60 strains were confirmed as members of the four recurrent lineages regardless of the method used (N = 32, 12, 7, and 9, respectively). Our results indicate that, regardless of the typing scheme used, epidemic or endemic signals were detected as reflected by lineage B (ST2254-gyrA9-porA1) in 2014 or lineage A (ST19-gyrA8-porA7), respectively. These findings support the clonal expansion of stable genomes in Campylobacter population exhibiting a multi-host profile and accounting for the majority of clinical strains isolated over a decade. Such recurring genotypes suggest persistence in reservoirs, sources or environment, emphasizing the need to investigate their survival strategy in greater depth.
Keywords: Campylobacter jejuni; WGS typing scheme comparison; clones; core genome MLST; recurring genotypes; typing schemes; whole genome MLST; whole genome sequencing.
Copyright © 2021 Nennig, Llarena, Herold, Mossong, Penny, Losch, Tresse and Ragimbeau.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Campylobacter MOMP database. Available at: https://pubmlst.org/campylobacter/info/porA_method.shtml (Accessed April 15, 2020).
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

