Accurate Detection of Target MicroRNA in Mixed Species of High Sequence Homology Using Target-Protection Rolling Circle Amplification
- PMID: 33490811
- PMCID: PMC7818630
- DOI: 10.1021/acsomega.0c05279
Accurate Detection of Target MicroRNA in Mixed Species of High Sequence Homology Using Target-Protection Rolling Circle Amplification
Abstract
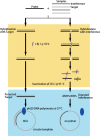
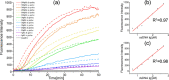
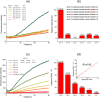
The close relationships of miRNAs with human diseases highlight the urgent needs for miRNA detection. However, the accurate detection of a target miRNA in mixed miRNAs of high sequence homology presents a great challenge. Herein, a novel method called target-protection rolling circle amplification (TP-RCA) is proposed for this purpose. The protective probe is designed so that it can form a fully complementary duplex with the target miRNA and can also mismatch duplexes with other nontarget miRNAs. These duplexes are treated with a single strand-specific nuclease. Consequently, only the target miRNA in a perfect-match duplex can resist the cleavage of nuclease, whereas the nontarget miRNAs in mismatched duplexes will be digested completely. The protected target miRNA can be detected using RCA reactions. MicroRNA let-7 family members (let-7a-let-7f) and nuclease CEL I were used as proof-of-concept models to evaluate the feasibility of the TP-RCA method under different experimental conditions. The experimental results show that the TP-RCA method can unambiguously detect the target let-7 species in mixtures of let-7 family members even though they may differ by only a single nucleotide. This TP-RCA method significantly improves the detection specificity of miRNAs.
© 2021 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

References
-
- Xu H.; Zhang Y.; Zhang S.; Sun M.; Li W.; Jiang Y.; Wu Z.-S. Ultrasensitive assay based on a combined cascade amplification by nicking-mediated rolling circle amplification and symmetric strand-displacement amplification. Anal. Chim. Acta 2019, 1047, 172–178. 10.1016/j.aca.2018.10.004. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

