The Impact of Intraspecies and Interspecies Bacterial Interactions on Disease Outcome
- PMID: 33494265
- PMCID: PMC7909810
- DOI: 10.3390/pathogens10020096
The Impact of Intraspecies and Interspecies Bacterial Interactions on Disease Outcome
Abstract
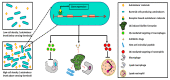
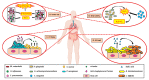
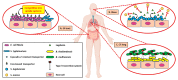
The human microbiota is an array of microorganisms known to interact with the host and other microbes. These interactions can be competitive, as microbes must adapt to host- and microorganism-related stressors, thus producing toxic molecules, or cooperative, whereby microbes survive by maintaining homeostasis with the host and host-associated microbial communities. As a result, these microbial interactions shape host health and can potentially result in disease. In this review, we discuss these varying interactions across microbial species, their positive and negative effects, the therapeutic potential of these interactions, and their implications on our knowledge of human well-being.
Keywords: bacterial interactions; commensal; host–pathogen interface; pathogen; polymicrobial infection.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Selective pressures during chronic infection drive microbial competition and cooperation.NPJ Biofilms Microbiomes. 2019 Jun 7;5(1):16. doi: 10.1038/s41522-019-0089-2. eCollection 2019. NPJ Biofilms Microbiomes. 2019. PMID: 31263568 Free PMC article. Review.
-
Polymicrobial Interactions Operative during Pathogen Transmission.mBio. 2021 May 18;12(3):e01027-21. doi: 10.1128/mBio.01027-21. mBio. 2021. PMID: 34006664 Free PMC article. Review.
-
From wolves to humans: oral microbiome resistance to transfer across mammalian hosts.mBio. 2024 Mar 13;15(3):e0334223. doi: 10.1128/mbio.03342-23. Epub 2024 Feb 1. mBio. 2024. PMID: 38299854 Free PMC article.
-
Microbiota-host interactions shape ageing dynamics.Philos Trans R Soc Lond B Biol Sci. 2020 Sep 28;375(1808):20190596. doi: 10.1098/rstb.2019.0596. Epub 2020 Aug 10. Philos Trans R Soc Lond B Biol Sci. 2020. PMID: 32772667 Free PMC article. Review.
-
Chemical Mediators at the Bacterial-Fungal Interface.Annu Rev Microbiol. 2020 Sep 8;74:267-290. doi: 10.1146/annurev-micro-012420-081224. Epub 2020 Jul 13. Annu Rev Microbiol. 2020. PMID: 32660387 Review.
Cited by
-
Rescue of pyrimidine-defective Pseudomonas aeruginosa through metabolic complementation.Microbiol Spectr. 2024 Jul 11;12(8):e0422623. doi: 10.1128/spectrum.04226-23. Online ahead of print. Microbiol Spectr. 2024. PMID: 38990029 Free PMC article.
-
Intraspecific cooperation allows the survival of Staphylococcus aureus staff: a novel strategy for disease relapse.BMC Infect Dis. 2024 Oct 1;24(1):1092. doi: 10.1186/s12879-024-10001-2. BMC Infect Dis. 2024. PMID: 39354412 Free PMC article.
-
Heritable Tissue-Specific Gene Expression Associates With Chronic Wound Microbial Species.Wound Repair Regen. 2025 Jul-Aug;33(4):e70055. doi: 10.1111/wrr.70055. Wound Repair Regen. 2025. PMID: 40576243 Free PMC article.
-
Development of a Polymicrobial Checkerboard Assay as a Tool for Determining Combinatorial Antibiotic Effectiveness in Polymicrobial Communities.Antibiotics (Basel). 2023 Jul 20;12(7):1207. doi: 10.3390/antibiotics12071207. Antibiotics (Basel). 2023. PMID: 37508303 Free PMC article.
-
Perspectives in Searching Antimicrobial Peptides (AMPs) Produced by the Microbiota.Microb Ecol. 2023 Dec 1;87(1):8. doi: 10.1007/s00248-023-02313-8. Microb Ecol. 2023. PMID: 38036921 Free PMC article. Review.
References
-
- Fons A.G.M. Mechanisms of Colonisation and Colonisation Resistance of the Digestive Tract Part 2: Bacteria/Bacteria Interactions. Microb. Ecol. Health Dis. 2000;12:240–246. doi: 10.1080/089106000750060495. - DOI
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

