Misincorporation Proteomics Technologies: A Review
- PMID: 33494504
- PMCID: PMC7924376
- DOI: 10.3390/proteomes9010002
Misincorporation Proteomics Technologies: A Review
Erratum in
-
Correction: Steele et al. Misincorporation Proteomics Technologies: A Review. Proteomes 2021, 9, 2.Proteomes. 2022 Jun 16;10(2):22. doi: 10.3390/proteomes10020022. Proteomes. 2022. PMID: 35736802 Free PMC article.
Abstract
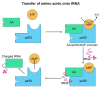
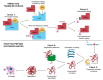
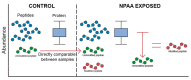
Proteinopathies are diseases caused by factors that affect proteoform conformation. As such, a prevalent hypothesis is that the misincorporation of noncanonical amino acids into a proteoform results in detrimental structures. However, this hypothesis is missing proteomic evidence, specifically the detection of a noncanonical amino acid in a peptide sequence. This review aims to outline the current state of technology that can be used to investigate mistranslations and misincorporations whilst framing the pursuit as Misincorporation Proteomics (MiP). The current availability of technologies explored herein is mass spectrometry, sample enrichment/preparation, data analysis techniques, and the hyphenation of approaches. While many of these technologies show potential, our review reveals a need for further development and refinement of approaches is still required.
Keywords: misincorporation; non protein amino acids; post translational modifications.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

