Identification of downstream signaling cascades of ACK1 and prognostic classifiers in non-small cell lung cancer
- PMID: 33495411
- PMCID: PMC7906148
- DOI: 10.18632/aging.202408
Identification of downstream signaling cascades of ACK1 and prognostic classifiers in non-small cell lung cancer
Abstract
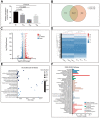
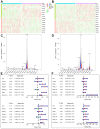
Activated Cdc42-associated kinase 1 (ACK1) is an oncogene in multiple cancers, but the underlying mechanisms of its oncogenic role remain unclear in non-small cell lung cancer (NSCLC). Herein, we comprehensively investigated the ACK1-regulated cell processes and downstream signaling pathways, as well as its prognostic value in NSCLC. We found that ACK1 gene amplification was associated with mRNA levels in The Cancer Genome Atlas (TCGA) lung cancer cohort. The Oncomine databases showed significantly elevated ACK1 levels in lung cancer. In vitro, an ACK1 inhibitor (dasatinib) increased the sensitivity of NSCLC cell lines to AKT or MEK inhibitors. RNA-sequencing results demonstrated that an ACK1 deficiency in A549 cells affected the MAPK, PI3K/AKT, and Wnt pathways. These results were validated by gene set enrichment analysis (GSEA) of data from 188 lung cancer cell lines. Using Cytoscape, we dissected 14 critical ACK1-regulated genes. The signature with the 14 genes and ACK1 could significantly dichotomize the TCGA lung cohort regarding overall survival. The prognostic accuracy of this signature was confirmed in five independent lung cancer cohorts and was further validated by a prognostic nomogram. Our study unveiled several downstream signaling pathways for ACK1, and the proposed signature may be a promising prognostic predictor for NSCLC.
Keywords: ACK1; NSCLC; TCGA; dasatinib; prognosis.
Conflict of interest statement
Figures







References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

