Whole genome sequencing of metastatic colorectal cancer reveals prior treatment effects and specific metastasis features
- PMID: 33495476
- PMCID: PMC7835235
- DOI: 10.1038/s41467-020-20887-6
Whole genome sequencing of metastatic colorectal cancer reveals prior treatment effects and specific metastasis features
Erratum in
-
Author Correction: Whole genome sequencing of metastatic colorectal cancer reveals prior treatment effects and specific metastasis features.Nat Commun. 2021 May 26;12(1):3269. doi: 10.1038/s41467-021-23629-4. Nat Commun. 2021. PMID: 34039979 Free PMC article. No abstract available.
Abstract
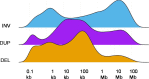
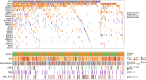
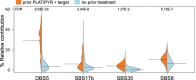
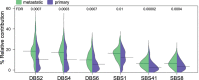
In contrast to primary colorectal cancer (CRC) little is known about the genomic landscape of metastasized CRC. Here we present whole genome sequencing data of metastases of 429 CRC patients participating in the pan-cancer CPCT-02 study (NCT01855477). Unsupervised clustering using mutational signature patterns highlights three major patient groups characterized by signatures known from primary CRC, signatures associated with received prior treatments, and metastasis-specific signatures. Compared to primary CRC, we identify additional putative (non-coding) driver genes and increased frequencies in driver gene mutations. In addition, we identify specific genes preferentially affected by microsatellite instability. CRC-specific 1kb-10Mb deletions, enriched for common fragile sites, and LINC00672 mutations are associated with response to treatment in general, whereas FBXW7 mutations predict poor response specifically to EGFR-targeted treatment. In conclusion, the genomic landscape of mCRC shows defined changes compared to primary CRC, is affected by prior treatments and contains features with potential clinical relevance.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

