Bloom syndrome DNA helicase deficiency is associated with oxidative stress and mitochondrial network changes
- PMID: 33495511
- PMCID: PMC7835382
- DOI: 10.1038/s41598-021-81075-0
Bloom syndrome DNA helicase deficiency is associated with oxidative stress and mitochondrial network changes
Abstract
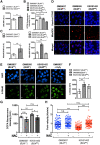
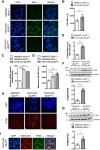
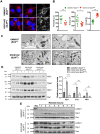
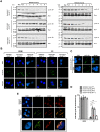
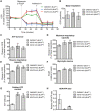
Bloom Syndrome (BS; OMIM #210900; ORPHA #125) is a rare genetic disorder that is associated with growth deficits, compromised immune system, insulin resistance, genome instability and extraordinary predisposition to cancer. Most efforts thus far have focused on understanding the role of the Bloom syndrome DNA helicase BLM as a recombination factor in maintaining genome stability and suppressing cancer. Here, we observed increased levels of reactive oxygen species (ROS) and DNA base damage in BLM-deficient cells, as well as oxidative-stress-dependent reduction in DNA replication speed. BLM-deficient cells exhibited increased mitochondrial mass, upregulation of mitochondrial transcription factor A (TFAM), higher ATP levels and increased respiratory reserve capacity. Cyclin B1, which acts in complex with cyclin-dependent kinase CDK1 to regulate mitotic entry and associated mitochondrial fission by phosphorylating mitochondrial fission protein Drp1, fails to be fully degraded in BLM-deficient cells and shows unscheduled expression in G1 phase cells. This failure to degrade cyclin B1 is accompanied by increased levels and persistent activation of Drp1 throughout mitosis and into G1 phase as well as mitochondrial fragmentation. This study identifies mitochondria-associated abnormalities in Bloom syndrome patient-derived and BLM-knockout cells and we discuss how these abnormalities may contribute to Bloom syndrome.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Endogenous gamma-H2AX-ATM-Chk2 checkpoint activation in Bloom's syndrome helicase deficient cells is related to DNA replication arrested forks.Mol Cancer Res. 2007 Jul;5(7):713-24. doi: 10.1158/1541-7786.MCR-07-0028. Mol Cancer Res. 2007. PMID: 17634426
-
Augmented cell death with Bloom syndrome helicase deficiency.Mol Med Rep. 2011 Jul-Aug;4(4):607-9. doi: 10.3892/mmr.2011.484. Epub 2011 May 3. Mol Med Rep. 2011. PMID: 21567087
-
Bloom syndrome cells undergo p53-dependent apoptosis and delayed assembly of BRCA1 and NBS1 repair complexes at stalled replication forks.J Cell Biol. 2003 Sep 29;162(7):1197-209. doi: 10.1083/jcb.200304016. J Cell Biol. 2003. PMID: 14517203 Free PMC article.
-
Oxidative stress, mitochondrial abnormalities and antioxidant defense in Ataxia-telangiectasia, Bloom syndrome and Nijmegen breakage syndrome.Redox Biol. 2017 Apr;11:375-383. doi: 10.1016/j.redox.2016.12.030. Epub 2016 Dec 28. Redox Biol. 2017. PMID: 28063379 Free PMC article. Review.
-
Bloom syndrome and the underlying causes of genetic instability.Mol Genet Metab. 2021 May;133(1):35-48. doi: 10.1016/j.ymgme.2021.03.003. Epub 2021 Mar 10. Mol Genet Metab. 2021. PMID: 33736941 Review.
Cited by
-
Cistanche deserticola polysaccharides alleviate cognitive decline in aging model mice by restoring the gut microbiota-brain axis.Aging (Albany NY). 2021 Jun 3;13(11):15320-15335. doi: 10.18632/aging.203090. Epub 2021 Jun 3. Aging (Albany NY). 2021. PMID: 34081627 Free PMC article.
-
BLM helicase overexpressed in human gliomas contributes to diverse responses of human glioma cells to chemotherapy.Cell Death Discov. 2023 May 11;9(1):157. doi: 10.1038/s41420-023-01451-9. Cell Death Discov. 2023. PMID: 37169803 Free PMC article.
-
Condensin II interacts with BLM helicase in S phase to maintain genome stability.Commun Biol. 2025 Mar 25;8(1):492. doi: 10.1038/s42003-025-07916-0. Commun Biol. 2025. PMID: 40133469 Free PMC article.
-
Mitochondrial superoxide dismutase Sod2 suppresses nuclear genome instability during oxidative stress.Genetics. 2023 Oct 4;225(2):iyad147. doi: 10.1093/genetics/iyad147. Genetics. 2023. PMID: 37638880 Free PMC article.
-
Severe Insulin Resistance Syndromes: Clinical Spectrum and Management.Int J Mol Sci. 2025 Jun 13;26(12):5669. doi: 10.3390/ijms26125669. Int J Mol Sci. 2025. PMID: 40565151 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

