Shared genetic pathways contribute to risk of hypertrophic and dilated cardiomyopathies with opposite directions of effect
- PMID: 33495596
- PMCID: PMC7611259
- DOI: 10.1038/s41588-020-00762-2
Shared genetic pathways contribute to risk of hypertrophic and dilated cardiomyopathies with opposite directions of effect
Abstract
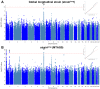
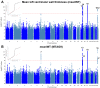
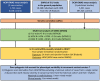
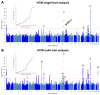
The heart muscle diseases hypertrophic (HCM) and dilated (DCM) cardiomyopathies are leading causes of sudden death and heart failure in young, otherwise healthy, individuals. We conducted genome-wide association studies and multi-trait analyses in HCM (1,733 cases), DCM (5,521 cases) and nine left ventricular (LV) traits (19,260 UK Biobank participants with structurally normal hearts). We identified 16 loci associated with HCM, 13 with DCM and 23 with LV traits. We show strong genetic correlations between LV traits and cardiomyopathies, with opposing effects in HCM and DCM. Two-sample Mendelian randomization supports a causal association linking increased LV contractility with HCM risk. A polygenic risk score explains a significant portion of phenotypic variability in carriers of HCM-causing rare variants. Our findings thus provide evidence that polygenic risk score may account for variability in Mendelian diseases. More broadly, we provide insights into how genetic pathways may lead to distinct disorders through opposing genetic effects.
Conflict of interest statement
M.-P.D. is author on a patent pertaining to pharmacogenomics-guided CETP inhibition (US20170233812A1), has a minor equity interest in DalCor and has received honoraria from Dalcor and Servier and research support (access to samples and data) from AstraZeneca, Pfizer, Servier, Sanofi and GlaxoSmithKline. J.-C.T. has received research grants from Amarin, AstraZeneca, DalCor, Esperion, Ionis, Sanofi and Servier; honoraria from AstraZeneca, DalCor, HLS, Sanofi and Servier; holds minor equity interest in DalCor; and is an author of a patent on pharmacogenomics-guided CETP inhibition (US20170233812A1). B.M. has received Research Funding from Siemens Healtheneers, Daiichi Sankyo. The UMCG, which employs R.A.d.B., has received research grants and/or fees from AstraZeneca, Abbott, Bristol-Myers Squibb, Novartis, Novo Nordisk, and Roche. R.A.d.B. received speaker fees from Abbott, AstraZeneca, Novartis, and Roche. HW is a consultant for Cytokinetics. P.M.M. receives an honorarium as Chair of the UKRI Medical Research Council Neuroscience and Mental Health Board. He acknowledges consultancy fees from Adelphi Communications, MedScape, Neurodiem, Nodthera, Biogen, Celgene and Roche. He has received speakers’ honoraria from Celgene, Biogen, Novartis and Roche, and has received research or educational funds from Biogen, GlaxoSmithKline and Novartis. He is paid as a member of the Scientific Advisory Board for Ipsen Pharmaceuticals. J.S.W. has received research support and consultancy fees from Myokardia, Inc.
Figures
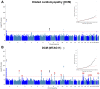
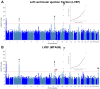
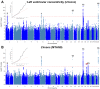
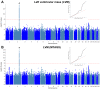
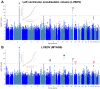
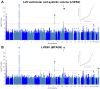
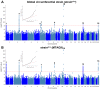
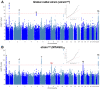
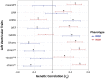
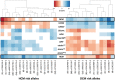
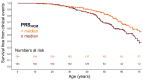
Comment in
-
New insights into the genetics of cardiomyopathies.Nat Rev Cardiol. 2021 Apr;18(4):229. doi: 10.1038/s41569-021-00526-3. Nat Rev Cardiol. 2021. PMID: 33564142 No abstract available.
References
-
- Semsarian C, Ingles J, Maron MS, Maron BJ. New perspectives on the prevalence of hypertrophic cardiomyopathy. J Am Coll Cardiol. 2015;65:1249–1254. - PubMed
-
- Elliott PM, et al. 2014 ESC Guidelines on diagnosis and management of hypertrophic cardiomyopathy: the Task Force for the Diagnosis and Management of Hypertrophic Cardiomyopathy of the European Society of Cardiology (ESC) Eur Heart J. 2014;35:2733–2779. - PubMed
Publication types
MeSH terms
Grants and funding
- MC_UP_1102/19/MRC_/Medical Research Council/United Kingdom
- G0601966/MRC_/Medical Research Council/United Kingdom
- 107469/Z/15/Z/WT_/Wellcome Trust/United Kingdom
- RG/19/6/34387/BHF_/British Heart Foundation/United Kingdom
- MC_UP_1102/20/MRC_/Medical Research Council/United Kingdom
- U01 HL117006/HL/NHLBI NIH HHS/United States
- 090532/Z/09/Z/WT_/Wellcome Trust/United Kingdom
- RE/18/4/34215/BHF_/British Heart Foundation/United Kingdom
- SP/17/11/32885/BHF_/British Heart Foundation/United Kingdom
- MR/K501013/1/MRC_/Medical Research Council/United Kingdom
- MR/S003754/1/MRC_/Medical Research Council/United Kingdom
- NH/17/1/32725/BHF_/British Heart Foundation/United Kingdom
- MC_U120085815/MRC_/Medical Research Council/United Kingdom
- 107469/WT_/Wellcome Trust/United Kingdom
- MR/N026934/1/MRC_/Medical Research Council/United Kingdom
- RG/18/9/33887/BHF_/British Heart Foundation/United Kingdom
- 91815610/ZONMW_/ZonMw/Netherlands
- MC_PC_17114/MRC_/Medical Research Council/United Kingdom
- FS/15/81/31817/BHF_/British Heart Foundation/United Kingdom
- DH_/Department of Health/United Kingdom
- 087183/WT_/Wellcome Trust/United Kingdom
- G0300665/MRC_/Medical Research Council/United Kingdom
- MC_UP_1605/13/MRC_/Medical Research Council/United Kingdom
- MR/M024903/1/MRC_/Medical Research Council/United Kingdom
- G9901399/MRC_/Medical Research Council/United Kingdom
- G9409531/MRC_/Medical Research Council/United Kingdom
- G0900897/MRC_/Medical Research Council/United Kingdom
- G9409634/MRC_/Medical Research Council/United Kingdom
- G0100811/MRC_/Medical Research Council/United Kingdom
- RE/13/1/30181/BHF_/British Heart Foundation/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources

