Hepatitis C Virus Transmission Clusters in Public Health and Correctional Settings, Wisconsin, USA, 2016-20171
- PMID: 33496239
- PMCID: PMC7853590
- DOI: 10.3201/eid2702.202957
Hepatitis C Virus Transmission Clusters in Public Health and Correctional Settings, Wisconsin, USA, 2016-20171
Abstract
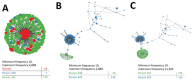
Ending the hepatitis C virus (HCV) epidemic requires stopping transmission among networks of persons who inject drugs. Identifying transmission networks by using genomic epidemiology may inform community responses that can quickly interrupt transmission. We retrospectively identified HCV RNA-positive specimens corresponding to 459 persons in settings that use the state laboratory, including correctional facilities and syringe services programs, in Wisconsin, USA, during 2016-2017. We conducted next-generation sequencing of HCV and analyzed it for phylogenetic linkage by using the Centers for Disease Control and Prevention Global Hepatitis Outbreak Surveillance Technology platform. Analysis showed that 126 persons were linked across 42 clusters. Phylogenetic clustering was higher in rural communities and associated with female sex and younger age among rural residents. These data highlight that HCV transmission could be reduced by expanding molecular-based surveillance strategies to rural communities affected by the opioid crisis.
Keywords: Hepatitis C virus; United States; Wisconsin; global hepatitis outbreak surveillance technology; hepatitis; injection drug use; molecular epidemiology; phylogenetics; transmission clusters; viruses.
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

