Outbreak of Severe Vomiting in Dogs Associated with a Canine Enteric Coronavirus, United Kingdom
- PMID: 33496240
- PMCID: PMC7853541
- DOI: 10.3201/eid2702.202452
Outbreak of Severe Vomiting in Dogs Associated with a Canine Enteric Coronavirus, United Kingdom
Abstract
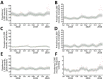
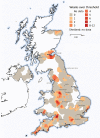
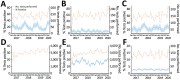
The lack of population health surveillance for companion animal populations leaves them vulnerable to the effects of novel diseases without means of early detection. We present evidence on the effectiveness of a system that enabled early detection and rapid response a canine gastroenteritis outbreak in the United Kingdom. In January 2020, prolific vomiting among dogs was sporadically reported in the United Kingdom. Electronic health records from a nationwide sentinel network of veterinary practices confirmed a significant increase in dogs with signs of gastroenteric disease. Male dogs and dogs living with other vomiting dogs were more likely to be affected. Diet and vaccination status were not associated with the disease; however, a canine enteric coronavirus was significantly associated with illness. The system we describe potentially fills a gap in surveillance in neglected populations and could provide a blueprint for other countries.
Keywords: United Kingdom; canine enteric coronavirus; dogs; enteric infections; gastrointestinal disease; outbreaks; statistical modeling; surveillance; syndromic surveillance; viruses; vomiting.
Figures

Similar articles
-
Small animal disease surveillance 2020/21: SARS-CoV-2, syndromic surveillance and an outbreak of acute vomiting in UK dogs.Vet Rec. 2021 Apr;188(8):304. doi: 10.1002/vetr.427. Vet Rec. 2021. PMID: 33891730 Free PMC article. No abstract available.
-
Emerging Variants of Canine Enteric Coronavirus Associated with Outbreaks of Gastroenteric Disease.Emerg Infect Dis. 2024 Jun;30(6):1240-1244. doi: 10.3201/eid3006.231184. Emerg Infect Dis. 2024. PMID: 38782018 Free PMC article.
-
Identification of a canine coronavirus in Australian racing Greyhounds.J Vet Diagn Invest. 2022 Jan;34(1):77-81. doi: 10.1177/10406387211054819. Epub 2021 Oct 26. J Vet Diagn Invest. 2022. PMID: 34697969 Free PMC article.
-
Canine respiratory coronavirus: an emerging pathogen in the canine infectious respiratory disease complex.Vet Clin North Am Small Anim Pract. 2008 Jul;38(4):815-25, viii. doi: 10.1016/j.cvsm.2008.02.008. Vet Clin North Am Small Anim Pract. 2008. PMID: 18501280 Free PMC article. Review.
-
Canine coronavirus: not only an enteric pathogen.Vet Clin North Am Small Anim Pract. 2011 Nov;41(6):1121-32. doi: 10.1016/j.cvsm.2011.07.005. Epub 2011 Sep 29. Vet Clin North Am Small Anim Pract. 2011. PMID: 22041207 Free PMC article. Review.
Cited by
-
An investigation into an outbreak of pancytopenia in cats in the United Kingdom.J Vet Intern Med. 2023 Jan;37(1):117-125. doi: 10.1111/jvim.16615. Epub 2023 Jan 7. J Vet Intern Med. 2023. PMID: 36610017 Free PMC article.
-
Small animal disease surveillance 2020/21: SARS-CoV-2, syndromic surveillance and an outbreak of acute vomiting in UK dogs.Vet Rec. 2021 Apr;188(8):304. doi: 10.1002/vetr.427. Vet Rec. 2021. PMID: 33891730 Free PMC article. No abstract available.
-
PetBERT: automated ICD-11 syndromic disease coding for outbreak detection in first opinion veterinary electronic health records.Sci Rep. 2023 Oct 21;13(1):18015. doi: 10.1038/s41598-023-45155-7. Sci Rep. 2023. PMID: 37865683 Free PMC article.
-
Using topic modelling for unsupervised annotation of electronic health records to identify an outbreak of disease in UK dogs.PLoS One. 2021 Dec 9;16(12):e0260402. doi: 10.1371/journal.pone.0260402. eCollection 2021. PLoS One. 2021. PMID: 34882714 Free PMC article.
-
Coronaviruses in wild animals sampled in and around Wuhan at the beginning of COVID-19 emergence.Virus Evol. 2022 Jun 4;8(1):veac046. doi: 10.1093/ve/veac046. eCollection 2022. Virus Evol. 2022. PMID: 35769892 Free PMC article.
References
-
- Smith S, Elliot AJ, Mallaghan C, Modha D, Hippisley-Cox J, Large S, et al. Value of syndromic surveillance in monitoring a focal waterborne outbreak due to an unusual Cryptosporidium genotype in Northamptonshire, United Kingdom, June - July 2008. Euro Surveill. 2010;15:19643. 10.2807/ese.15.33.19643-en - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

