Using Open Data to Rapidly Benchmark Biomolecular Simulations: Phospholipid Conformational Dynamics
- PMID: 33496579
- PMCID: PMC7903423
- DOI: 10.1021/acs.jcim.0c01299
Using Open Data to Rapidly Benchmark Biomolecular Simulations: Phospholipid Conformational Dynamics
Abstract
Molecular dynamics (MD) simulations are widely used to monitor time-resolved motions of biomacromolecules, although it often remains unknown how closely the conformational dynamics correspond to those occurring in real life. Here, we used a large set of open-access MD trajectories of phosphatidylcholine (PC) lipid bilayers to benchmark the conformational dynamics in several contemporary MD models (force fields) against nuclear magnetic resonance (NMR) data available in the literature: effective correlation times and spin-lattice relaxation rates. We found none of the tested MD models to fully reproduce the conformational dynamics. That said, the dynamics in CHARMM36 and Slipids are more realistic than in the Amber Lipid14, OPLS-based MacRog, and GROMOS-based Berger force fields, whose sampling of the glycerol backbone conformations is too slow. The performance of CHARMM36 persists when cholesterol is added to the bilayer, and when the hydration level is reduced. However, for conformational dynamics of the PC headgroup, both with and without cholesterol, Slipids provides the most realistic description because CHARMM36 overestimates the relative weight of ∼1 ns processes in the headgroup dynamics. We stress that not a single new simulation was run for the present work. This demonstrates the worth of open-access MD trajectory databanks for the indispensable step of any serious MD study: benchmarking the available force fields. We believe this proof of principle will inspire other novel applications of MD trajectory databanks and thus aid in developing biomolecular MD simulations into a true computational microscope-not only for lipid membranes but for all biomacromolecular systems.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
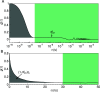
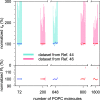
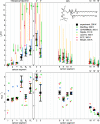



References
-
- Crystallography: Protein Data Bank. Nat. New Biol. 1971, 233, 223.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

