Specifications of the variant curation guidelines for ITGA2B/ITGB3: ClinGen Platelet Disorder Variant Curation Panel
- PMID: 33496739
- PMCID: PMC7839359
- DOI: 10.1182/bloodadvances.2020003712
Specifications of the variant curation guidelines for ITGA2B/ITGB3: ClinGen Platelet Disorder Variant Curation Panel
Abstract
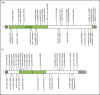
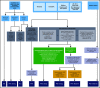
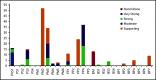
Accurate and consistent sequence variant interpretation is critical to the correct diagnosis and appropriate clinical management and counseling of patients with inherited genetic disorders. To minimize discrepancies in variant curation and classification among different clinical laboratories, the American College of Medical Genetics and Genomics (ACMG), along with the Association for Molecular Pathology (AMP), published standards and guidelines for the interpretation of sequence variants in 2015. Because the rules are not universally applicable to different genes or disorders, the Clinical Genome Resource (ClinGen) Platelet Disorder Expert Panel (PD-EP) has been tasked to make ACMG/AMP rule specifications for inherited platelet disorders. ITGA2B and ITGB3, the genes underlying autosomal recessive Glanzmann thrombasthenia (GT), were selected as the pilot genes for specification. Eight types of evidence covering clinical phenotype, functional data, and computational/population data were evaluated in the context of GT by the ClinGen PD-EP. The preliminary specifications were validated with 70 pilot ITGA2B/ITGB3 variants and further refined. In the final adapted criteria, gene- or disease-based specifications were made to 16 rules, including 7 with adjustable strength; no modification was made to 5 rules; and 7 rules were deemed not applicable to GT. Employing the GT-specific ACMG/AMP criteria to the pilot variants resulted in a reduction of variants classified with unknown significance from 29% to 20%. The overall concordance with the initial expert assertions was 71%. These adapted criteria will serve as guidelines for GT-related variant interpretation to increase specificity and consistency across laboratories and allow for better clinical integration of genetic knowledge into patient care.
Conflict of interest statement
Conflict-of-interest disclosure: B.R.B., S.N.D., M.L., and J.P.B. work for clinical diagnostic laboratories that offer testing for GT. B.M.Z. is involved in the curation and interpretation of exome sequencing in patients with inherited platelet disorders, as well as the development of an NGS diagnostic gene panel for inherited platelet disorder. M.P.L. is an advisory board member for Octapharma and Shionogi, is a consultant for Amgen, Novartis, Shionogi, Dova, Principia, Argenx, Rigel, and Bayer, and has received research funding from Sysmex, Novartis, Rigel, and AstraZeneca. The remaining authors declare no competing financial interests.
Figures



Similar articles
-
Specifications of the ACMG/AMP variant curation guidelines for myocilin: Recommendations from the clingen glaucoma expert panel.Hum Mutat. 2022 Dec;43(12):2170-2186. doi: 10.1002/humu.24482. Epub 2022 Oct 19. Hum Mutat. 2022. PMID: 36217948 Free PMC article.
-
ClinGen Myeloid Malignancy Variant Curation Expert Panel recommendations for germline RUNX1 variants.Blood Adv. 2019 Oct 22;3(20):2962-2979. doi: 10.1182/bloodadvances.2019000644. Blood Adv. 2019. PMID: 31648317 Free PMC article.
-
Variant Classification for Pompe disease; ACMG/AMP specifications from the ClinGen Lysosomal Diseases Variant Curation Expert Panel.Mol Genet Metab. 2023 Sep-Oct;140(1-2):107715. doi: 10.1016/j.ymgme.2023.107715. Epub 2023 Oct 26. Mol Genet Metab. 2023. PMID: 37907381 Free PMC article.
-
Glanzmann thrombasthenia: a review of ITGA2B and ITGB3 defects with emphasis on variants, phenotypic variability, and mouse models.Blood. 2011 Dec 1;118(23):5996-6005. doi: 10.1182/blood-2011-07-365635. Epub 2011 Sep 13. Blood. 2011. PMID: 21917754 Review.
-
Understanding the genetic basis of Glanzmann thrombasthenia: implications for treatment.Expert Rev Hematol. 2012 Oct;5(5):487-503. doi: 10.1586/ehm.12.46. Expert Rev Hematol. 2012. PMID: 23146053 Review.
Cited by
-
The effects of pathogenic and likely pathogenic variants for inherited hemostasis disorders in 140 214 UK Biobank participants.Blood. 2023 Dec 14;142(24):2055-2068. doi: 10.1182/blood.2023020118. Blood. 2023. PMID: 37647632 Free PMC article.
-
GoldVariants, a resource for sharing rare genetic variants detected in bleeding, thrombotic, and platelet disorders: Communication from the ISTH SSC Subcommittee on Genomics in Thrombosis and Hemostasis.J Thromb Haemost. 2021 Oct;19(10):2612-2617. doi: 10.1111/jth.15459. Epub 2021 Aug 5. J Thromb Haemost. 2021. PMID: 34355501 Free PMC article.
-
Genetics of inherited thrombocytopenias.Blood. 2022 Jun 2;139(22):3264-3277. doi: 10.1182/blood.2020009300. Blood. 2022. PMID: 35167650 Free PMC article. Review.
-
Hemostatic phenotypes and genetic disorders.Res Pract Thromb Haemost. 2021 Dec 16;5(8):e12637. doi: 10.1002/rth2.12637. eCollection 2021 Dec. Res Pract Thromb Haemost. 2021. PMID: 34964017 Free PMC article.
-
Canadian College of Medical Geneticists: clinical practice advisory document - responsibility to recontact for reinterpretation of clinical genetic testing.J Med Genet. 2024 Nov 25;61(12):1123-1131. doi: 10.1136/jmg-2024-110330. J Med Genet. 2024. PMID: 39362754 Free PMC article.
References
-
- Amendola LM, Jarvik GP, Leo MC, et al. . Performance of ACMG-AMP variant-interpretation guidelines among nine laboratories in the Clinical Sequencing Exploratory Research Consortium [published correction appears in Am J Hum Genet. 2016;99(1):247]. Am J Hum Genet. 2016;98(6):1067-1076. - PMC - PubMed
-
- Richards S, Aziz N, Bale S, et al. ; ACMG Laboratory Quality Assurance Committee . Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405-424. - PMC - PubMed
-
- Rehm HL, Berg JS, Plon SE. ClinGen and ClinVar—enabling genomics in precision medicine. Hum Mutat. 2018;39(11):1473-1475.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

